7Y0V
 
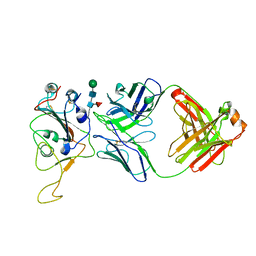 | | The co-crystal structure of BA.1-RBD with Fab-5549 | | Descriptor: | 5549-Fab, Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Xiao, J.Y, Zhang, Y. | | Deposit date: | 2022-06-06 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
7Y0C
 
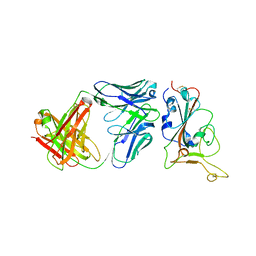 | | Crystal structure of BD55-1403 and SARS-CoV-2 Omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD55-1403 Fab heavy chain, BD55-1403 Fab light chain, ... | | Authors: | Zhang, Z, Xiao, J. | | Deposit date: | 2022-06-04 | | Release date: | 2022-09-28 | | Last modified: | 2023-04-12 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
5XRP
 
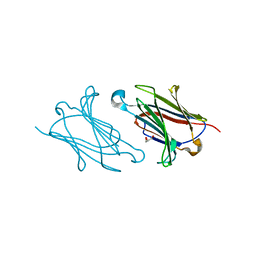 | | Galectin-10/Charcot-Leyden crystal protein variant Q75A | | Descriptor: | GLYCEROL, Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2017-06-08 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Galectin-10: a new structural type of prototype galectin dimer and effects on saccharide ligand binding.
Glycobiology, 28, 2018
|
|
5IO1
 
 | |
8I4T
 
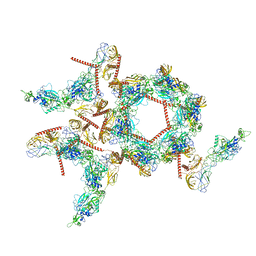 | | Structure of the asymmetric unit of SFTSV virion | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-01-21 | | Release date: | 2023-09-13 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
8ILQ
 
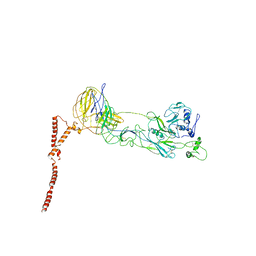 | | Structure of SFTSV Gn-Gc heterodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelopment polyprotein | | Authors: | Du, S, Peng, R, Qi, J, Li, C. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of severe fever with thrombocytopenia syndrome virus.
Nat Commun, 14, 2023
|
|
6JCJ
 
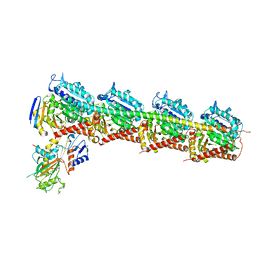 | | Structure of crolibulin in complex with tubulin | | Descriptor: | (4R)-2,7,8-triamino-4-(3-bromo-4,5-dimethoxyphenyl)-4H-1-benzopyran-3-carbonitrile, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Zhang, Z, Yang, J. | | Deposit date: | 2019-01-29 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism of crolibulin in complex with tubulin provides a rationale for drug design.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | Deposit date: | 2023-07-17 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
7EKQ
 
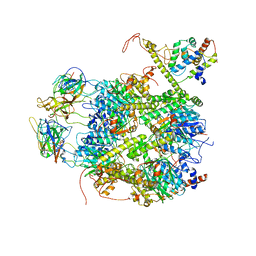 | | CrClpP-S2c | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit, ... | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
7EKO
 
 | | CrClpP-S1 | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit CLPT4, chloroplastic, ATP-dependent Clp protease proteolytic subunit | | Authors: | Wang, N, Wang, Y.F, Cong, Y, Liu, C.M. | | Deposit date: | 2021-04-06 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The cryo-EM structure of the chloroplast ClpP complex.
Nat.Plants, 7, 2021
|
|
8K3K
 
 | |
5XRO
 
 | |
5XRI
 
 | |
5XRJ
 
 | |
5XSU
 
 | | novel orally efficacious inhibitors complexed with PARP1 | | Descriptor: | 6-fluoranyl-2-(4,5,6,7-tetrahydrofuro[2,3-c]pyridin-2-yl)-1~{H}-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Liu, Q, Xu, Y. | | Deposit date: | 2017-06-15 | | Release date: | 2018-04-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of 2-(4,5,6,7-tetrahydrothienopyridin-2-yl)-benzoimidazole carboxamides as novel orally efficacious Poly(ADP-ribose)polymerase (PARP) inhibitors
Eur J Med Chem, 145, 2018
|
|
5XST
 
 | | novel orally efficacious inhibitors complexed with PARP1 | | Descriptor: | 6-fluoranyl-2-(4,5,6,7-tetrahydrothieno[3,2-c]pyridin-2-yl)-1~{H}-benzimidazole-4-carboxamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Liu, Q, Xu, Y. | | Deposit date: | 2017-06-15 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and synthesis of 2-(4,5,6,7-tetrahydrothienopyridin-2-yl)-benzoimidazole carboxamides as novel orally efficacious Poly(ADP-ribose)polymerase (PARP) inhibitors
Eur J Med Chem, 145, 2018
|
|
5XRL
 
 | |
4J1G
 
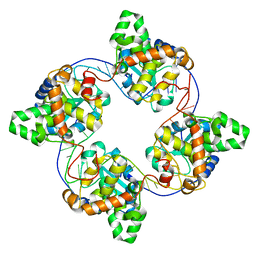 | | Leanyer orthobunyavirus nucleoprotein-ssRNA complex | | Descriptor: | Nucleocapsid, RNA (45-MER) | | Authors: | Niu, F, Shaw, N, Wang, Y, Jiao, L, Ding, W, Li, X, Zhu, P, Upur, H, Ouyang, S, Cheng, G, Liu, Z.J. | | Deposit date: | 2013-02-01 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Structure of the Leanyer orthobunyavirus nucleoprotein-RNA complex reveals unique architecture for RNA encapsidation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8EZ7
 
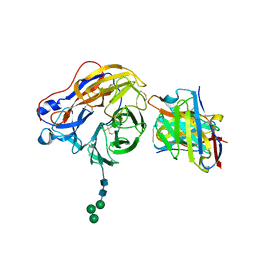 | | Structure of 1F04 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 1F04, Light chain of influenza virus neuraminidase antibody 1F04, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
8EZ8
 
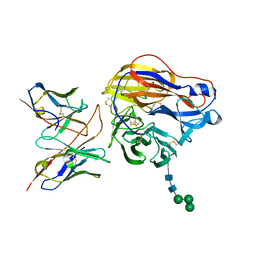 | | Structure of 3C08 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 3C08, Light chain of influenza virus neuraminidase antibody 3C08, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
8EZ3
 
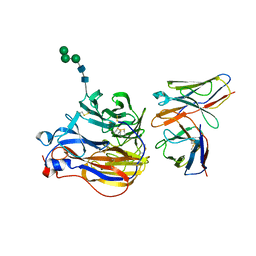 | | Structure of 3A10 Fab in complex with A/Moscow/10/1999 (H3N2) influenza virus neuraminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of influenza virus neuraminidase antibody 3A10, Light chain of influenza virus neuraminidase antibody 3A10, ... | | Authors: | Mou, Z, Lei, R, Wu, N.C, Dai, X. | | Deposit date: | 2022-10-31 | | Release date: | 2023-11-08 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Leveraging vaccination-induced protective antibodies to define conserved epitopes on influenza N2 neuraminidase.
Immunity, 56, 2023
|
|
8ETN
 
 | |
8FXU
 
 | | Thermoanaerobacter thermosaccharolyticum periplasmic Glucose-Binding Protein glucose complex: Badan conjugate attached at F17C | | Descriptor: | 2-bromo-1-[6-(dimethylamino)naphthalen-2-yl]ethan-1-one, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Allert, M.J, Kumar, S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Chromophore carbonyl twisting in fluorescent biosensors encodes direct readout of protein conformations with multicolor switching.
Commun Chem, 6, 2023
|
|
