4PBH
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM RUEGERIA POMEROYI DSS-3 (SPO1773, TARGET EFI-510260) WITH BOUND BENZOIC ACID | | Descriptor: | BENZOIC ACID, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-12 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PF6
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSEOBACTER DENITRIFICANS (RD1_0742, TARGET EFI-510239) WITH BOUND 3-DEOXY-D-MANNO-OCT-2-ULOSONIC ACID (KDO) | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, C4-dicarboxylate-binding protein, SULFATE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3P8O
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with des-bromine analogue of BI 201335 | | Descriptor: | HCV non-structural protein 4A, HCV serine protease NS3, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-N-[(1R,2S)-1-carboxy-2-ethenylcyclopropyl]-4-[(7-methoxy-2-{2-[(2-methylpropanoyl)amino]-1,3-thiazol-4-yl}quinolin-4-yl)oxy]-L-prolinamide, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2010-10-14 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Combined X-ray, NMR, and kinetic analyses reveal uncommon binding characteristics of the hepatitis C virus NS3-NS4A protease inhibitor BI 201335.
J.Biol.Chem., 286, 2011
|
|
4PGN
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM DESULFOVIBRIO ALASKENSIS G20 (Dde_0634, TARGET EFI-510120) WITH BOUND INDOLE PYRUVATE | | Descriptor: | 1,2-ETHANEDIOL, 3-(1H-INDOL-3-YL)-2-OXOPROPANOIC ACID, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PDD
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM POLAROMONAS SP JS666 (Bpro_0088, TARGET EFI-510167) BOUND TO D-ERYTHRONATE | | Descriptor: | (2R,3R)-2,3,4-trihydroxybutanoic acid, FORMIC ACID, MALONIC ACID, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PF8
 
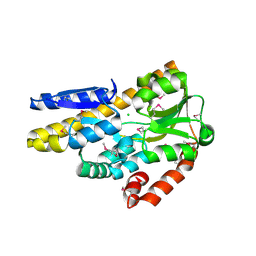 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM SULFITOBACTER sp. NAS-14.1 (TARGET EFI-510299) WITH BOUND BETA-D-GALACTURONATE | | Descriptor: | CHLORIDE ION, TRAP-T family transporter, DctP (Periplasmic binding) subunit, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PFB
 
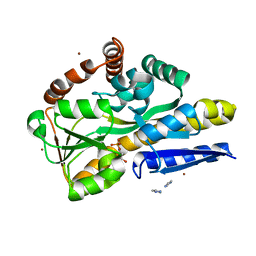 | | Crystal structure of a TRAP periplasmic solute binding protein from Fusobacterium nucleatun (FN1258, TARGET EFI-510120) with bound SN-glycerol-3-phosphate | | Descriptor: | C4-dicarboxylate-binding protein, IMIDAZOLE, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PET
 
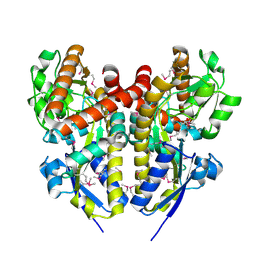 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM COLWELLIA PSYCHRERYTHRAEA (CPS_0129, TARGET EFI-510097) WITH BOUND CALCIUM AND PYRUVATE | | Descriptor: | CALCIUM ION, CHLORIDE ION, Extracellular solute-binding protein, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PC9
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM ROSENBACTER DENITRIFICANS OCh 114 (RD1_1052, TARGET EFI-510238) WITH BOUND D-MANNONATE | | Descriptor: | C4-dicarboxylate transport system, substrate-binding protein, putative, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-14 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PDH
 
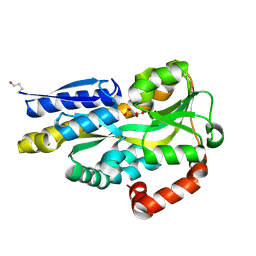 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM POLAROMONAS SP JS666 (Bpro_1871, TARGET EFI-510164) BOUND TO D-ERYTHRONATE | | Descriptor: | (2R,3R)-2,3,4-trihydroxybutanoic acid, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4PE3
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM RHODOBACTER SPHAEROIDES (Rsph17029_3620, TARGET EFI-510199), APO OPEN STRUCTURE | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, IMIDAZOLE, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-22 | | Release date: | 2014-05-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4P9K
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM VERMINEPHROBACTER EISENIAE EF01-2 (Veis_3954, TARGET EFI-510324) A NEPHRIDIAL SYMBIONT OF THE EARTHWORM EISENIA FOETIDA, BOUND TO D-ERYTHRONATE WITH RESIDUAL DENSITY SUGGESTIVE OF SUPERPOSITION WITH COPURIFIED ALTERNATIVE LIGAND. | | Descriptor: | (2R,3R)-2,3,4-trihydroxybutanoic acid, TRAP dicarboxylate transporter, DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-04 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N6D
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens DSM2638 (Desal_3247), Target EFI-510112, phased with I3C, open complex, C-terminus of symmetry mate bound in ligand binding site | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3P8N
 
 | | Crystal structure of HCV NS3/NS4A protease complexed with BI 201335 | | Descriptor: | HCV non-structural protein 4A, HCV serine protease NS3, N-[(cyclopentyloxy)carbonyl]-3-methyl-L-valyl-(4R)-4-[(8-bromo-7-methoxy-2-{2-[(2-methylpropanoyl)amino]-1,3-thiazol-4-yl}quinolin-4-yl)oxy]-N-[(1R,2S)-1-carboxy-2-ethenylcyclopropyl]-L-prolinamide, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2010-10-14 | | Release date: | 2011-01-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Combined X-ray, NMR, and kinetic analyses reveal uncommon binding characteristics of the hepatitis C virus NS3-NS4A protease inhibitor BI 201335.
J.Biol.Chem., 286, 2011
|
|
5GXU
 
 | | Cystal structure of Arabidopsis ATR2 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADPH--cytochrome P450 reductase 2 | | Authors: | Niu, G, Liu, L. | | Deposit date: | 2016-09-20 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Arabidopsis thaliana NADPH-cytochrome P450 reductase 2 (ATR2) provides insight into its function
FEBS J., 284, 2017
|
|
5XJ7
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the acyl phosphate form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION, ... | | Authors: | Tang, Y, Li, Z, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5XJ9
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the orthophosphate form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Glycerol-3-phosphate acyltransferase, PHOSPHATE ION | | Authors: | Li, Z, Tang, Y, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
5XJ5
 
 | | Crystal structure of PlsY (YgiH), an integral membrane glycerol 3-phosphate acyltransferase - the monoacylglycerol form | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, GLYCINE, Glycerol-3-phosphate acyltransferase, ... | | Authors: | Li, Z, Li, D. | | Deposit date: | 2017-04-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Structural insights into the committed step of bacterial phospholipid biosynthesis.
Nat Commun, 8, 2017
|
|
4DHF
 
 | | Structure of Aurora A mutant bound to Biogenidec cpd 15 | | Descriptor: | 7-cyclopentyl-2-({1-methyl-5-[(4-methylpiperazin-1-yl)carbonyl]-1H-pyrrol-3-yl}amino)-7H-pyrrolo[2,3-d]pyrimidine-6-carboxamide, Aurora kinase A, MAGNESIUM ION, ... | | Authors: | Silvian, L, Marcotte, D.J. | | Deposit date: | 2012-01-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of 2,6,7-trisubstituted-7H-pyrrolo[2,3-d]pyrimidines as Aurora kinases inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6XUE
 
 | |
6XUG
 
 | |
6XUQ
 
 | |
4XFR
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Bordetella bronchiseptica (BB3215), Target EFI-511620, with bound citrate, domain swapped dimer, space group P6522 | | Descriptor: | CITRIC ACID, Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-28 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XGJ
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Pectobacterium atrosepticum (ECA3761), Target EFI-511609, APO structure, domain swapped dimer | | Descriptor: | Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-30 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4XFM
 
 | | Crystal structure of a domain of unknown function (DUF1537) from Pectobacterium atrosepticum (ECA3761), Target EFI-511609, with bound D-threonate, domain swapped dimer | | Descriptor: | THREONATE ION, Uncharacterized protein | | Authors: | Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-12-27 | | Release date: | 2015-04-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Assignment of function to a domain of unknown function: DUF1537 is a new kinase family in catabolic pathways for acid sugars.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
