6Q16
 
 | | Focussed refinement of InvGN0N1:PrgHK:SpaPQR:PrgIJ from Salmonella SPI-1 injectisome NC-base | | Descriptor: | Lipoprotein PrgK, Protein InvG, Protein PrgH, ... | | Authors: | Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2019-08-02 | | Release date: | 2019-10-23 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | T3S injectisome needle complex structures in four distinct states reveal the basis of membrane coupling and assembly.
Nat Microbiol, 4, 2019
|
|
6QD9
 
 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000065 | | Descriptor: | 3-[[2-[3-(dimethylamino)propyl-methyl-amino]thieno[3,2-d]pyrimidin-4-yl]-methyl-amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
6QDH
 
 | | Leishmania major N-myristoyltransferase in complex with quinazoline inhibitor IMP-0000906 | | Descriptor: | 4-[ethyl(methyl)amino]-2-[methyl-(1-methylpiperidin-4-yl)amino]-~{N}-(1,3,5-trimethylpyrazol-4-yl)quinazoline-6-sulfonamide, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
6QDE
 
 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000877 | | Descriptor: | 3-[[6-~{tert}-butyl-2-[3-(dimethylamino)propyl-methyl-amino]thieno[3,2-d]pyrimidin-4-yl]-methyl-amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
6QDF
 
 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000096 | | Descriptor: | 3-[[6-tert-butyl-2-[methyl-[(3S)-1-methylpyrrolidin-3-yl]amino]thieno[3,2-d]pyrimidin-4-yl]-methyl-amino]propanenitrile, Glycylpeptide N-tetradecanoyltransferase, MAGNESIUM ION, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
6QDC
 
 | |
6QDD
 
 | | Leishmania major N-myristoyltransferase in complex with thienopyrimidine inhibitor IMP-0000105 | | Descriptor: | 2-[(2~{S})-1-[2-[methyl-(1-methylpiperidin-4-yl)amino]thieno[3,2-d]pyrimidin-4-yl]-2,3-dihydropyrrol-2-yl]ethanenitrile, DIMETHYL SULFOXIDE, Glycylpeptide N-tetradecanoyltransferase, ... | | Authors: | Brannigan, J.A. | | Deposit date: | 2019-01-01 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Novel Thienopyrimidine Inhibitors of Leishmania N -Myristoyltransferase with On-Target Activity in Intracellular Amastigotes.
J.Med.Chem., 63, 2020
|
|
8SBE
 
 | | Structure of the rat vesicular glutamate transporter 2 determined by single-particle Cryo-EM | | Descriptor: | Vesicular glutamate transporter 2 | | Authors: | Li, F, Finer-Moore, J, Eriksen, J, Cheng, Y, Edwards, R, Stroud, R. | | Deposit date: | 2023-04-03 | | Release date: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Ion transport and regulation in a synaptic vesicle glutamate transporter.
Science, 368, 2020
|
|
3N2Y
 
 | |
7JN4
 
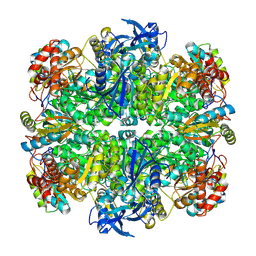 | | Rubisco in the apo state | | Descriptor: | Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain 2, chloroplastic | | Authors: | Matthies, D, Jonikas, M.C, He, S. | | Deposit date: | 2020-08-03 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | The structural basis of Rubisco phase separation in the pyrenoid.
Nat.Plants, 6, 2020
|
|
7JSX
 
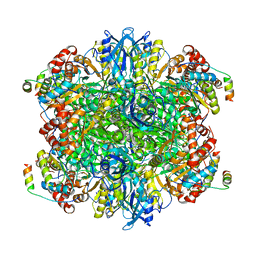 | | EPYC1(106-135) peptide-bound Rubisco | | Descriptor: | EPYC1, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain 2, ... | | Authors: | Matthies, D, He, S, Jonikas, M.C. | | Deposit date: | 2020-08-16 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.06 Å) | | Cite: | The structural basis of Rubisco phase separation in the pyrenoid.
Nat.Plants, 6, 2020
|
|
7JFO
 
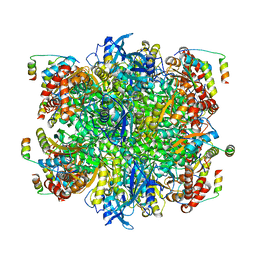 | | EPYC1(49-72)-bound Rubisco | | Descriptor: | LCI5, Ribulose bisphosphate carboxylase large chain, Ribulose bisphosphate carboxylase small chain 2, ... | | Authors: | Matthies, D, Jonikas, M.C, He, S. | | Deposit date: | 2020-07-17 | | Release date: | 2020-11-18 | | Last modified: | 2020-12-23 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | The structural basis of Rubisco phase separation in the pyrenoid.
Nat.Plants, 6, 2020
|
|
7LFS
 
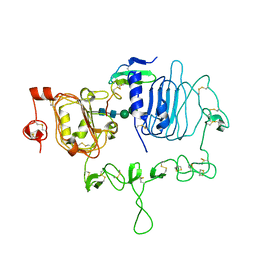 | | Crystal structure of the epidermal growth factor receptor extracellular region with A265V mutation in complex with epiregulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 4 of Epidermal growth factor receptor, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LEN
 
 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with trehalose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-14 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
7LFR
 
 | | Crystal structure of the epidermal growth factor receptor extracellular region with R84K mutation in complex with epiregulin crystallized with spermine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Proepiregulin, ... | | Authors: | Hu, C, Leche II, C.A, Stayrook, S.E, Ferguson, K.M, Lemmon, M.A. | | Deposit date: | 2021-01-18 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Glioblastoma mutations alter EGFR dimer structure to prevent ligand bias.
Nature, 602, 2022
|
|
8H1T
 
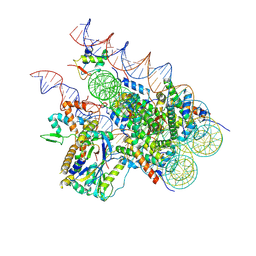 | | Cryo-EM structure of BAP1-ASXL1 bound to chromatosome | | Descriptor: | DNA (187-MER), Histone H1.4, Histone H2A type 1-D, ... | | Authors: | Ge, W, Yu, C, Xu, R.M. | | Deposit date: | 2022-10-04 | | Release date: | 2023-02-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase.
Nature, 616, 2023
|
|
7N9F
 
 | | Structure of the in situ yeast NPC | | Descriptor: | Dynein light chain 1, cytoplasmic, Nucleoporin 145c, ... | | Authors: | Villa, E, Singh, D, Ludtke, S.J, Akey, C.W, Rout, M.P, Echeverria, I, Suslov, S. | | Deposit date: | 2021-06-17 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N84
 
 | | Double nuclear outer ring from the isolated yeast NPC | | Descriptor: | Nucleoporin 145c, Nucleoporin NUP120, Nucleoporin NUP133, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7N85
 
 | | Inner ring spoke from the isolated yeast NPC | | Descriptor: | Nucleoporin ASM4, Nucleoporin NIC96, Nucleoporin NSP1, ... | | Authors: | Akey, C.W, Rout, M.P, Ouch, C, Echevarria, I, Fernandez-Martinez, J, Nudelman, I. | | Deposit date: | 2021-06-13 | | Release date: | 2022-01-26 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Comprehensive structure and functional adaptations of the yeast nuclear pore complex.
Cell, 185, 2022
|
|
7Y5U
 
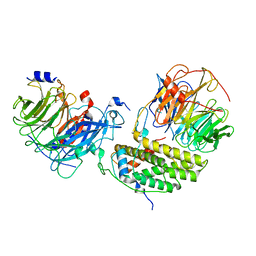 | | Cryo-EM structure of the monomeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5W
 
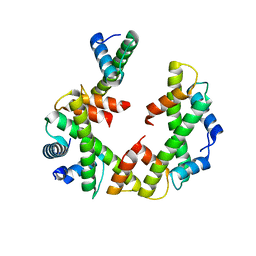 | | Cryo-EM structure of the left-handed Di-tetrasome | | Descriptor: | Histone H3.1, Histone H4, Widom 601 DNA (147-MER) | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y60
 
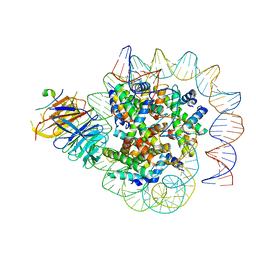 | | Cryo-EM structure of human CAF1LC bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y61
 
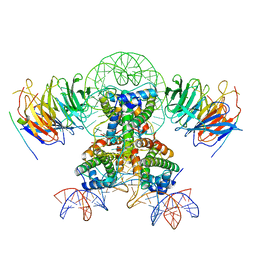 | | Cryo-EM structure of the two CAF1LCs bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Yu, C, Xu, R.M. | | Deposit date: | 2022-06-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7Y5V
 
 | | Cryo-EM structure of the dimeric human CAF1LC-H3-H4 complex | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, C, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2022-06-17 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
3RIK
 
 | | The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease | | Descriptor: | (3S,4R,5R,6S)-1-(2-hydroxyethyl)azepane-3,4,5,6-tetrol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | Authors: | Orwig, S.D, Lieberman, R.L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-03-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Binding of 3,4,5,6-tetrahydroxyazepanes to the acid-beta-glucosidase active site: implications for pharmacological chaperone design for Gaucher disease
Biochemistry, 50, 2011
|
|
