6LCY
 
 | | Crystal structure of Synaptotagmin-7 C2B in complex with IP6 | | Descriptor: | INOSITOL HEXAKISPHOSPHATE, Synaptotagmin-7 | | Authors: | Zhang, Y, Zhang, X, Rao, F, Wang, C. | | Deposit date: | 2019-11-20 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 5-IP 7 is a GPCR messenger mediating neural control of synaptotagmin-dependent insulin exocytosis and glucose homeostasis.
Nat Metab, 3, 2021
|
|
1N9U
 
 | | Differences and Similarities in Solution Structures of Angiotensin I & II: Implication for Structure-Function Relationship | | Descriptor: | Angiotensin I | | Authors: | Spyroulias, G.A, Nikolakopoulou, P, Tzakos, A, Gerothanassis, I.P, Magafa, V, Manessi-Zoupa, E, Cordopatis, P. | | Deposit date: | 2002-11-26 | | Release date: | 2003-07-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of angiotensin I & II. Implication for structure-function relationship.
Eur.J.Biochem., 270, 2003
|
|
1N9V
 
 | | Differences and Similarities in Solution Structures of Angiotensin I & II: Implication for Structure-Function Relationship. | | Descriptor: | Angiotensin II | | Authors: | Spyroulias, G.A, Nikolakopoulou, P, Tzakos, A, Gerothanassis, I.P, Magafa, V, Manessi-Zoupa, E, Cordopatis, P. | | Deposit date: | 2002-11-26 | | Release date: | 2003-07-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Comparison of the solution structures of angiotensin I & II. Implication for structure-function relationship.
Eur.J.Biochem., 270, 2003
|
|
7Y9G
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 in complex with pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7Y9H
 
 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diterpene synthase VenA, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7WZC
 
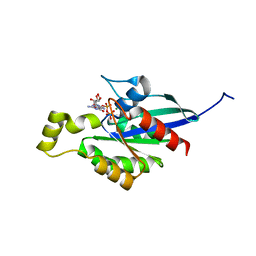 | | An open conformation Form2 of switch II for RhoA GDP-bound state | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79944921 Å) | | Cite: | A RhoA structure with switch II flipped outward revealed the conformational dynamics of switch II region.
J.Struct.Biol., 215, 2023
|
|
7WZA
 
 | | An open conformation Form 1 of switch II for RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Transforming protein RhoA | | Authors: | Jiang, H, Luo, C. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.50028777 Å) | | Cite: | A RhoA structure with switch II flipped outward revealed the conformational dynamics of switch II region.
J.Struct.Biol., 215, 2023
|
|
8Y59
 
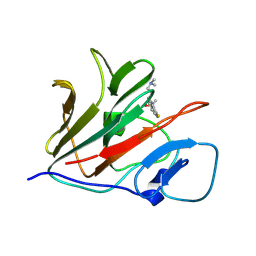 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with (S)-hydroxyl-acepromazine. | | Descriptor: | (1~{S})-1-[10-[3-(dimethylamino)propyl]phenothiazin-2-yl]ethanol, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Selective degradation of multimeric proteins via chemically induced proximity to TRIM21.
To be published
|
|
8Y5B
 
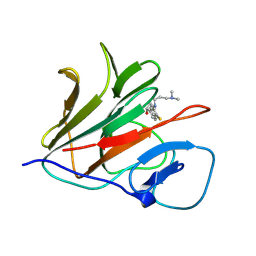 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with (R)-hydroxyl-acepromazine. | | Descriptor: | (1~{R})-1-[10-[3-(dimethylamino)propyl]phenothiazin-2-yl]ethanol, E3 ubiquitin-protein ligase TRIM21 | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Selective degradation of multimeric proteins via chemically induced proximity to TRIM21.
To be published
|
|
8Y58
 
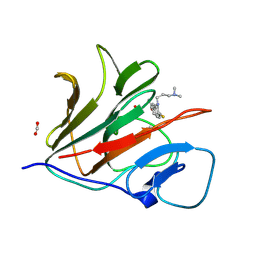 | | Crystal structure of TRIM21 PRYSPRY (D355A) in complex with acepromazine. | | Descriptor: | 1-[10-(3-DIMETHYLAMINO-PROPYL)-10H-PHENOTHIAZIN-2-YL]-ETHANONE, E3 ubiquitin-protein ligase TRIM21, FORMIC ACID | | Authors: | Lu, P, Cheng, Y, Xue, L, Ren, X, Huang, N, Han, T. | | Deposit date: | 2024-01-31 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective degradation of multimeric proteins via chemically induced proximity to TRIM21.
To be published
|
|
8Y74
 
 | |
7XN4
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with NAD+ | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN5
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Arginine ADP-riboxanase CopC, Calmodulin-1, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN6
 
 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
5YEG
 
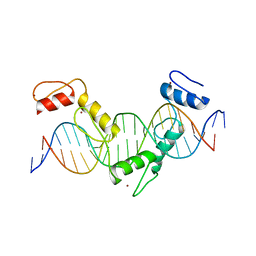 | | Crystal structure of CTCF ZFs4-8-Hs5-1a complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*AP*AP*CP*CP*AP*GP*CP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*GP*CP*TP*GP*GP*TP*TP*AP*AP*AP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Yin, M, Wang, J, Wang, M, Li, X. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
5YEL
 
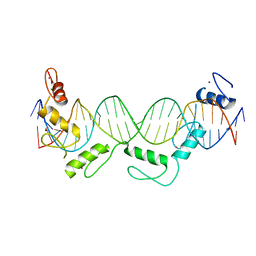 | | Crystal structure of CTCF ZFs6-11-gb7CSE | | Descriptor: | DNA (26-MER), Transcriptional repressor CTCF, ZINC ION | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-18 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
5WC2
 
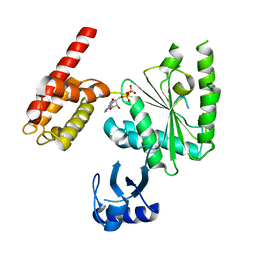 | | Crystal Structure of ADP-bound human TRIP13 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | Authors: | Jeong, B.-C, Luo, X. | | Deposit date: | 2017-06-29 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing.
Nat Commun, 8, 2017
|
|
5YEH
 
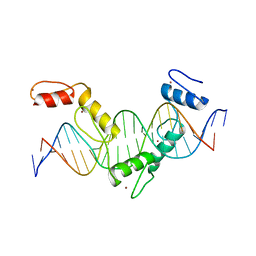 | | Crystal structure of CTCF ZFs4-8-eCBS | | Descriptor: | DNA (5'-D(*AP*CP*GP*GP*TP*TP*TP*CP*CP*GP*CP*TP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*AP*GP*CP*GP*GP*AP*AP*AP*CP*CP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.328 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
5YEF
 
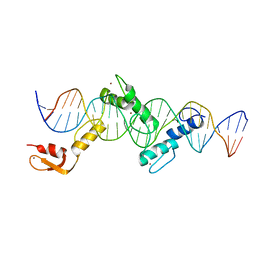 | | Crystal structure of CTCF ZFs2-8-Hs5-1aE | | Descriptor: | DNA (27-MER), Transcriptional repressor CTCF, ZINC ION | | Authors: | Yin, M, Wang, J, Wang, M, Li, X, Wang, Y. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.807 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
5Z2C
 
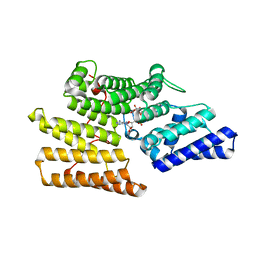 | | Crystal structure of ALPK-1 N-terminal domain in complex with ADP-heptose | | Descriptor: | Alpha-protein kinase 1, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2S,3S,4S,5S,6R)-6-[(1S)-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl] hydrogen phosphate | | Authors: | Ding, J, She, Y, Shao, F. | | Deposit date: | 2018-01-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Alpha-kinase 1 is a cytosolic innate immune receptor for bacterial ADP-heptose.
Nature, 561, 2018
|
|
7BTN
 
 | |
7CMO
 
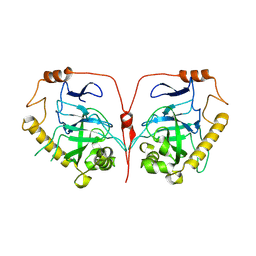 | | Crystal structure of human inorganic pyrophosphatase | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Hu, F, Huang, Z, Li, L. | | Deposit date: | 2020-07-28 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and biochemical characterization of inorganic pyrophosphatase from Homo sapiens.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7EG0
 
 | | Cryo-EM structure of anagrelide-induced PDE3A-SLFN12 complex | | Descriptor: | 6,7-bis(chloranyl)-3,5-dihydro-1H-imidazo[2,1-b]quinazolin-2-one, MAGNESIUM ION, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-23 | | Release date: | 2021-09-29 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG4
 
 | | Cryo-EM structure of nauclefine-induced PDE3A-SLFN12 complex | | Descriptor: | MAGNESIUM ION, Parvine, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-29 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
7EG1
 
 | | Cryo-EM structure of DNMDP-induced PDE3A-SLFN12 complex | | Descriptor: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, Schlafen family member 12, ... | | Authors: | Liu, N, Chen, J, Wang, X.D, Wang, H.W. | | Deposit date: | 2021-03-23 | | Release date: | 2021-11-03 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of PDE3A-SLFN12 complex and structure-based design for a potent apoptosis inducer of tumor cells.
Nat Commun, 12, 2021
|
|
