2ZVL
 
 | | Crystal structure of PCNA in complex with DNA polymerase kappa fragment | | Descriptor: | DNA polymerase kappa, Proliferating cell nuclear antigen, SULFATE ION, ... | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
2Z5M
 
 | | Complex of Transportin 1 with TAP NLS, crystal form 2 | | Descriptor: | Nuclear RNA export factor 1, Transportin-1 | | Authors: | Imasaki, T, Shimizu, T, Hashimoto, H, Hidaka, Y, Kose, S, Imamoto, N, Yamada, M, Sato, M. | | Deposit date: | 2007-07-14 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for substrate recognition and dissociation by human transportin 1
Mol.Cell, 28, 2007
|
|
2Z5O
 
 | | Complex of Transportin 1 with JKTBP NLS | | Descriptor: | Heterogeneous nuclear ribonucleoprotein D-like, Transportin-1 | | Authors: | Imasaki, T, Shimizu, T, Hashimoto, H, Hidaka, Y, Kose, S, Imamoto, N, Yamada, M, Sato, M. | | Deposit date: | 2007-07-14 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for substrate recognition and dissociation by human transportin 1
Mol.Cell, 28, 2007
|
|
3ABE
 
 | | Structure of human REV7 in complex with a human REV3 fragment in a tetragonal crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
3AUP
 
 | | Crystal structure of Basic 7S globulin from soybean | | Descriptor: | Basic 7S globulin | | Authors: | Yoshizawa, T, Shimizu, T, Taichi, M, Nishiuchi, Y, Yamabe, M, Shichijo, N, Unzai, S, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-02-14 | | Release date: | 2011-04-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Crystal structure of basic 7S globulin, a xyloglucan-specific endo-beta-1,4-glucanase inhibitor protein-like protein from soybean lacking inhibitory activity against endo-beta-glucanase
Febs J., 278, 2011
|
|
2Z5K
 
 | | Complex of Transportin 1 with TAP NLS | | Descriptor: | Nuclear RNA export factor 1, PHOSPHATE ION, Transportin-1 | | Authors: | Imasaki, T, Shimizu, T, Hashimoto, H, Hidaka, Y, Yamada, M, Kose, S, Imamoto, N, Sato, M. | | Deposit date: | 2007-07-14 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for substrate recognition and dissociation by human transportin 1
Mol.Cell, 28, 2007
|
|
3ABD
 
 | | Structure of human REV7 in complex with a human REV3 fragment in a monoclinic crystal | | Descriptor: | DNA polymerase zeta catalytic subunit, Mitotic spindle assembly checkpoint protein MAD2B | | Authors: | Hara, K, Hashimoto, H, Murakumo, Y, Kobayashi, S, Kogame, T, Unzai, S, Akashi, S, Takeda, S, Shimizu, T, Sato, M. | | Deposit date: | 2009-12-07 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of human REV7 in complex with a human REV3 fragment and structural implication of the interaction between DNA polymerase {zeta} and REV1
J.Biol.Chem., 285, 2010
|
|
2ZFU
 
 | | Structure of the methyltransferase-like domain of nucleomethylin | | Descriptor: | Cerebral protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Minami, H, Hashimoto, H, Murayama, A, Yanagisawa, J, Sato, M, Shimizu, T. | | Deposit date: | 2008-01-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Epigenetic control of rDNA loci in response to intracellular energy status
Cell(Cambridge,Mass.), 133, 2008
|
|
5YAQ
 
 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with scyllo-inosose | | Descriptor: | (2R,3S,4s,5R,6S)-2,3,4,5,6-pentahydroxycyclohexanone, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5YAB
 
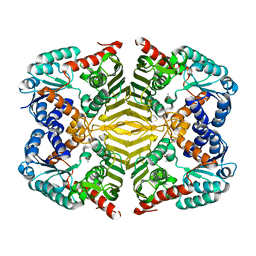 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Descriptor: | ACETATE ION, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-05-23 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5Y7Y
 
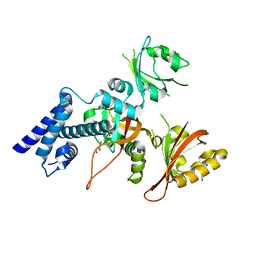 | | Crystal structure of AhRR/ARNT complex | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator, Aryl hydrocarbon receptor repressor, GLYCEROL | | Authors: | Sakurai, S, Shimizu, T, Ohto, U. | | Deposit date: | 2017-08-18 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of the AhRR-ARNT heterodimer reveals the structural basis of the repression of AhR-mediated transcription.
J. Biol. Chem., 292, 2017
|
|
5YAP
 
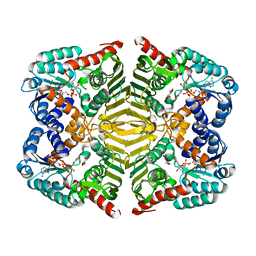 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with L-glucono-1,5-lactone | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-09-01 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5YA8
 
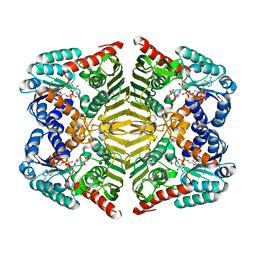 | | Crystal structure of scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity complexed with myo-inositol | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Fukano, K, Shimizu, T, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2017-08-31 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of L-glucose oxidation by scyllo-inositol dehydrogenase: Implications for a novel enzyme subfamily classification
PLoS ONE, 13, 2018
|
|
5Z15
 
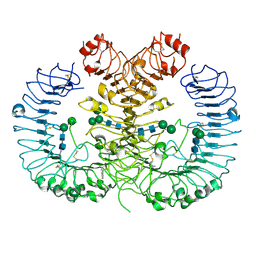 | | Crystal structure of human TLR8 in complex with CU-CPT9c | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(7-chloranylquinolin-4-yl)-2-methyl-phenol, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-12-25 | | Release date: | 2018-07-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of Toll-like Receptor 8 by Specifically Targeting a Unique Allosteric Site and Locking Its Resting State
To Be Published
|
|
5Z14
 
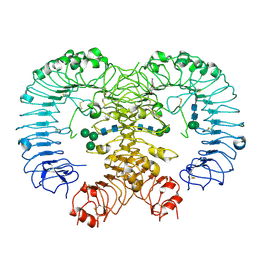 | | Crystal structure of human TLR8 in complex with CU-CPT9a | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(7-methoxyquinolin-4-yl)-2-methyl-phenol, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2017-12-25 | | Release date: | 2018-07-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inhibition of Toll-like Receptor 8 by Specifically Targeting a Unique Allosteric Site and Locking Its Resting State
To Be Published
|
|
5XDR
 
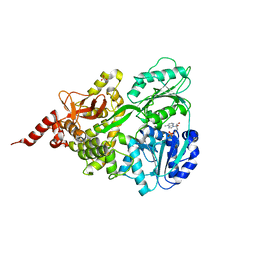 | | Crystal structure of human DEAH-box RNA helicase DHX15 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15, ... | | Authors: | Murakami, K, Nakano, K, Shimizu, T, Ohto, U. | | Deposit date: | 2017-03-29 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of human DEAH-box RNA helicase 15 reveals a domain organization of the mammalian DEAH/RHA family
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3VL8
 
 | | Crystal structure of XEG | | Descriptor: | SULFATE ION, Xyloglucan-specific endo-beta-1,4-glucanase A | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3VL9
 
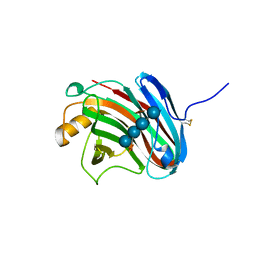 | | Crystal structure of xeg-xyloglucan | | Descriptor: | Xyloglucan-specific endo-beta-1,4-glucanase A, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3VLA
 
 | | Crystal structure of edgp | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EDGP | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3VLB
 
 | | Crystal structure of xeg-edgp | | Descriptor: | EDGP, Xyloglucan-specific endo-beta-1,4-glucanase A | | Authors: | Yoshizawa, T, Shimizu, T, Hirano, H, Sato, M, Hashimoto, H. | | Deposit date: | 2011-11-30 | | Release date: | 2012-04-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for inhibition of xyloglucan-specific endo-beta-1,4-glucanase (XEG) by XEG-protein inhibitor
J.Biol.Chem., 287, 2012
|
|
3VJH
 
 | | Human PPAR GAMMA ligand binding domain in complex with JKPL35 | | Descriptor: | (2S)-2-[4-methoxy-3-({[4-(trifluoromethyl)benzoyl]amino}methyl)benzyl]pentanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Tomioka, D, Kuwabara, N, Hashimoto, H, Sato, M, Shimizu, T. | | Deposit date: | 2011-10-20 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Peroxisome proliferator-activated receptors (PPARs) have multiple binding points that accommodate ligands in various conformations: phenylpropanoic acid-type PPAR ligands bind to PPAR in different conformations, depending on the subtype.
J.Med.Chem., 55, 2012
|
|
3THC
 
 | |
3THD
 
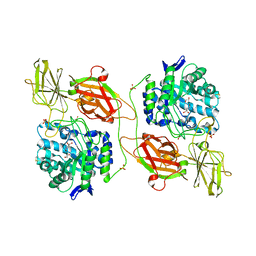 | | Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin | | Descriptor: | (2R,3S,4R,5S)-2-(hydroxymethyl)piperidine-3,4,5-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ohto, U, Shimizu, T. | | Deposit date: | 2011-08-18 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of human beta-galactosidase: structural basis of Gm1 gangliosidosis and morquio B diseases
J.Biol.Chem., 287, 2012
|
|
3W3G
 
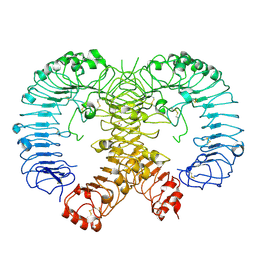 | | Crystal structure of human TLR8 (unliganded form) | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-21 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
3W3M
 
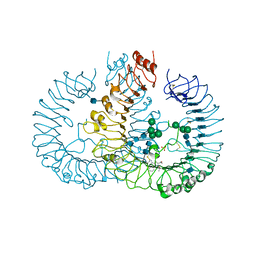 | | Crystal structure of human TLR8 in complex with Resiquimod (R848) crystal form 2 | | Descriptor: | 1-[4-amino-2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-1-yl]-2-methylpropan-2-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, SULFATE ION, ... | | Authors: | Tanji, H, Ohto, U, Shimizu, T. | | Deposit date: | 2012-12-22 | | Release date: | 2013-04-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural reorganization of the Toll-like receptor 8 dimer induced by agonistic ligands
Science, 339, 2013
|
|
