6L7U
 
 | | Crystal structure of FKRP in complex with Ba ion, Ba-SAD data | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BARIUM ION, Fukutin-related protein, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
6L7S
 
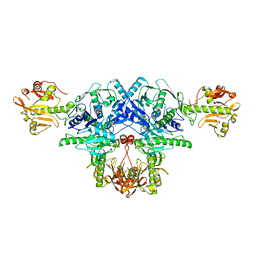 | | Crystal structure of FKRP in complex with Mg ion, Zinc peak data | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fukutin-related protein, MAGNESIUM ION, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
5AYF
 
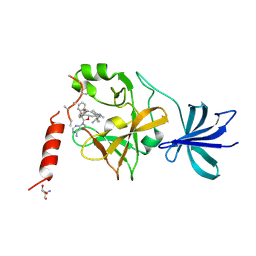 | | Crystal structure of SET7/9 in complex with cyproheptadine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(dibenzo[1,2-a:2',1'-d][7]annulen-11-ylidene)-1-methyl-piperidine, Histone-lysine N-methyltransferase SETD7, ... | | Authors: | Niwa, H, Handa, N, Takemoto, Y, Ito, A, Tomabechi, Y, Umehara, T, Shirouzu, M, Yoshida, M, Yokoyama, S. | | Deposit date: | 2015-08-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.005 Å) | | Cite: | Identification of Cyproheptadine as an Inhibitor of SET Domain Containing Lysine Methyltransferase 7/9 (Set7/9) That Regulates Estrogen-Dependent Transcription
J.Med.Chem., 59, 2016
|
|
6L7T
 
 | | Crystal structure of FKRP in complex with Mg ion, Zinc low remote data | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fukutin-related protein, MAGNESIUM ION, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2019-11-03 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structures of fukutin-related protein (FKRP), a ribitol-phosphate transferase related to muscular dystrophy.
Nat Commun, 11, 2020
|
|
5B08
 
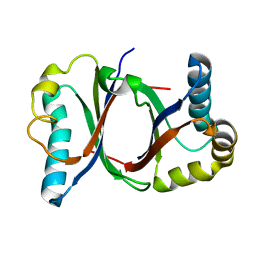 | | Polyketide cyclase OAC from Cannabis sativa | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.325 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0A
 
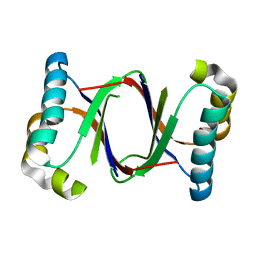 | | Polyketide cyclase OAC from Cannabis sativa, H5Q mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0E
 
 | | Polyketide cyclase OAC from Cannabis sativa, V59M mutant | | Descriptor: | GLYCEROL, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0F
 
 | | Polyketide cyclase OAC from Cannabis sativa, Y72F mutant | | Descriptor: | GLYCEROL, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0C
 
 | | Polyketide cyclase OAC from Cannabis sativa, Y27F mutant | | Descriptor: | Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
5B0B
 
 | | Polyketide cyclase OAC from Cannabis sativa, I7F mutant | | Descriptor: | ACETATE ION, Olivetolic acid cyclase | | Authors: | Yang, X, Matsui, T, Mori, T, Abe, I, Morita, H. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa
Febs J., 283, 2016
|
|
7DBN
 
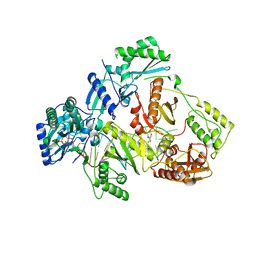 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V/F160M:DNA:dCTP ternary complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
7DBM
 
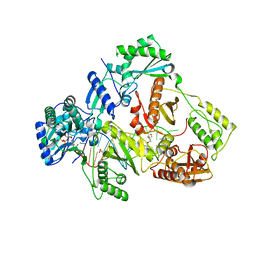 | | HIV-1 reverse transcriptase mutant Q151M/Y115F/F116Y/M184V:DNA:dGTP ternary complex | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA/RNA (38-MER), GLYCEROL, ... | | Authors: | Yasutake, Y, Hattori, S.I, Tamura, N, Maeda, K. | | Deposit date: | 2020-10-21 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Biochemical and Structural Properties of Entecavir-Resistant Hepatitis B Virus Polymerase with L180M/M204V Mutations.
J.Virol., 95, 2021
|
|
8HHQ
 
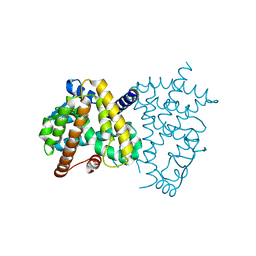 | |
8HHP
 
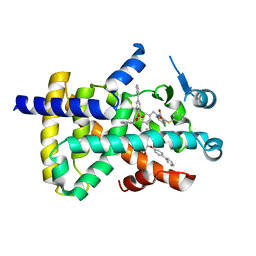 | |
8JJE
 
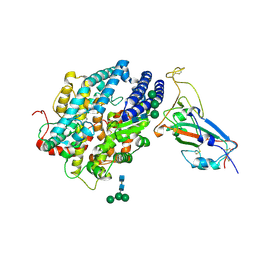 | | RBD of SARS-CoV2 spike protein with ACE2 decoy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Kishikawa, J, Hirose, M, Kato, T, Okamoto, T. | | Deposit date: | 2023-05-30 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An inhaled ACE2 decoy confers protection against SARS-CoV-2 infection in preclinical models.
Sci Transl Med, 15, 2023
|
|
8ZFW
 
 | |
9BR7
 
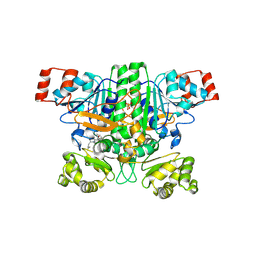 | | Crystal structure of human succinyl-CoA:glutarate-CoA transferase (SUGCT) in complex with Losartan carboxylic acid | | Descriptor: | AMMONIUM ION, SULFATE ION, Succinate--hydroxymethylglutarate CoA-transferase, ... | | Authors: | Wu, R, Lazarus, M.B. | | Deposit date: | 2024-05-10 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Characterization, Structure, and Inhibition of the Human Succinyl-CoA:glutarate-CoA Transferase, a Putative Genetic Modifier of Glutaric Aciduria Type 1.
Acs Chem.Biol., 19, 2024
|
|
9BR6
 
 | |
7P3I
 
 | | Crystal structure of human CD40/TNFRSF5 in complex with the anti-CD40 DARPin protein | | Descriptor: | Darpin, SODIUM ION, Tumor necrosis factor receptor superfamily member 5 | | Authors: | Malvezzi, F, Mangold, S, Hospodarsch, T, Reichen, C, Iss, C, Lammens, A, Krapp, S, Domke, C. | | Deposit date: | 2021-07-07 | | Release date: | 2022-04-06 | | Last modified: | 2022-05-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Multispecific Anti-CD40 DARPin Construct Induces Tumor-Selective CD40 Activation and Tumor Regression.
Cancer Immunol Res, 10, 2022
|
|
5GGI
 
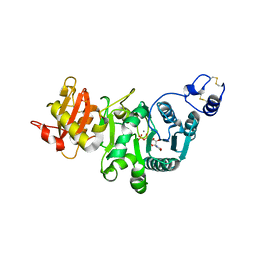 | | Crystal structure of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Mn, UDP and Mannosyl-peptide | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGK
 
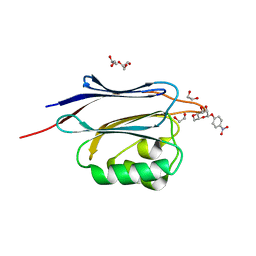 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with Man-beta-pNP | | Descriptor: | 1,2-ETHANEDIOL, 4-nitrophenyl beta-D-mannopyranoside, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGO
 
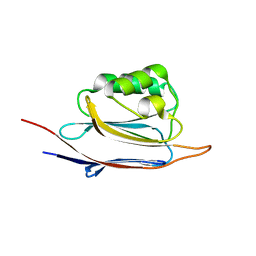 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GalNac-beta1,3-GlcNAc-beta-pNP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GGF
 
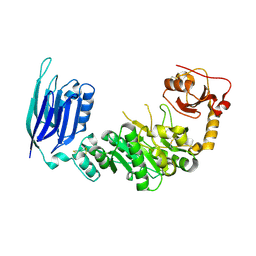 | |
5GGG
 
 | |
5GGN
 
 | | Crystal structure of N-terminal domain of human protein O-mannose beta-1,2-N-acetylglucosaminyltransferase in complex with GlcNAc-beta-pNP | | Descriptor: | 4-nitrophenyl 2-acetamido-2-deoxy-beta-D-glucopyranoside, Protein O-linked-mannose beta-1,2-N-acetylglucosaminyltransferase 1 | | Authors: | Kuwabara, N, Senda, T, Kato, R. | | Deposit date: | 2016-06-16 | | Release date: | 2016-08-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Carbohydrate-binding domain of the POMGnT1 stem region modulates O-mannosylation sites of alpha-dystroglycan
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
