8SGN
 
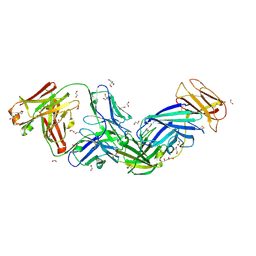 | | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy651H02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy651H02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine
To Be Published
|
|
8SGG
 
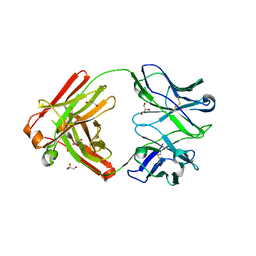 | | Crystal structure of Cy137D09, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine | | Descriptor: | Cy137D09 Fab heavy chain, Cy137D09 Fab light chain, GLYCEROL | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-12 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure of Cy137D09, a monoclonal antibody isolated from macaques immunized with an Epstein-Barr virus glycoprotein 350 (gp350) nanoparticle vaccine
To Be Published
|
|
8SIC
 
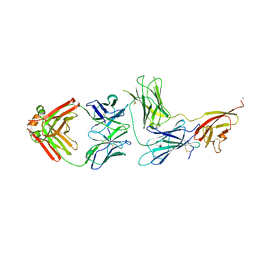 | | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy137C02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cy137C02 Fab heavy chain, Cy137C02 Fab light chain, ... | | Authors: | Joyce, M.G, Jensen, J.L, Chen, W.H, Kanekiyo, M. | | Deposit date: | 2023-04-14 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Crystal structure of Epstein-Barr virus glycoprotein 350 (gp350) in complex with Cy137C02, a monoclonal antibody isolated from macaques immunized with a gp350 nanoparticle vaccine
To Be Published
|
|
8SM1
 
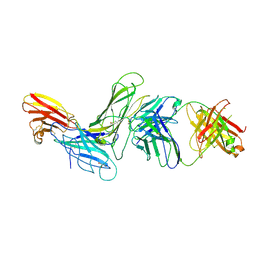 | | CRYSTAL STRUCTURE OF HUMAN ANTIBODY 769A9 IN COMPLEX WITH EPSTEIN-BARR VIRUS MAJOR GLYCOPROTEIN GP350 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769A9 Fab heavy chain, 769A9 Fab light chain, ... | | Authors: | Chen, W.-H, Bu, W, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural Basis For Receptor Engagement And Virus Neutralization Through Epstein-Barr Virus Gp350
To Be Published
|
|
8SM0
 
 | | Crystal structure of human complement receptor 2 (CD21) in complex with Epstein-Barr virus major glycoprotein gp350 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement receptor type 2, Envelope glycoprotein gp350, ... | | Authors: | Chen, W.-H, Bu, W, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2023-04-25 | | Release date: | 2024-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural Basis For Receptor Engagement And Virus Neutralization Through Epstein-Barr Virus Gp350
To Be Published
|
|
5BZ6
 
 | | Crystal structure of the N-terminal domain single mutant (S92A) of the human mitochondrial calcium uniporter fused with T4 lysozyme | | Descriptor: | Lysozyme,Calcium uniporter protein, mitochondrial, SULFATE ION | | Authors: | Lee, Y, Min, C.K, Kim, T.G, Song, H.K, Lim, Y, Kim, D, Shin, K, Kang, M, Kang, J.Y, Youn, H.-S, Lee, J.-G, An, J.Y, Park, K.R, Lim, J.J, Kim, J.H, Kim, J.H, Park, Z.Y, Kim, Y.-S, Wang, J, Kim, D.H, Eom, S.H. | | Deposit date: | 2015-06-11 | | Release date: | 2015-09-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and function of the N-terminal domain of the human mitochondrial calcium uniporter.
Embo Rep., 16, 2015
|
|
3S1I
 
 | | Crystal structure of oxygen-bound hell's gate globin I | | Descriptor: | Hemoglobin-like flavoprotein, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
3S1J
 
 | | Crystal structure of acetate-bound hell's gate globin I | | Descriptor: | ACETATE ION, Hemoglobin-like flavoprotein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Teh, A.H, Saito, J.A, Baharuddin, A, Tuckerman, J.R, Newhouse, J.S, Kanbe, M, Newhouse, E.I, Rahim, R.A, Favier, F, Didierjean, C, Sousa, E.H.S, Stott, M.B, Dunfield, P.F, Gonzalez, G, Gilles-Gonzalez, M.A, Najimudin, N, Alam, M. | | Deposit date: | 2011-05-15 | | Release date: | 2011-09-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hell's Gate globin I: an acid and thermostable bacterial hemoglobin resembling mammalian neuroglobin
Febs Lett., 585, 2011
|
|
1NP7
 
 | | Crystal Structure Analysis of Synechocystis sp. PCC6803 cryptochrome | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Brudler, R, Hitomi, K, Daiyasu, H, Toh, H, Kucho, K, Ishiura, M, Kanehisa, M, Roberts, V.A, Todo, T, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2003-01-17 | | Release date: | 2003-01-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a new cryptochrome class: structure, function, and evolution
Mol.Cell, 11, 2003
|
|
2J14
 
 | | 3,4,5-Trisubstituted Isoxazoles as Novel PPARdelta Agonists: Part2 | | Descriptor: | (3-{4-[2-(2,4-DICHLORO-PHENOXY)-ETHYLCARBAMOYL]-5-PHENYL-ISOXAZOL-3-YL}-PHENYL)-ACETIC ACID, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR DELTA | | Authors: | Epple, R, Azimioara, M, Russo, R, Xie, Y, Wang, X, Cow, C, Wityak, J, Karanewsky, D, Bursulaya, B, Kreusch, A, Tuntland, T, Gerken, A, Iskandar, M, Saez, E, Seidel, H.M, Tian, S.S. | | Deposit date: | 2006-08-08 | | Release date: | 2006-09-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3,4,5-Trisubstituted Isoxazoles as Novel Ppardelta Agonists. Part 2
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6FZC
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant L184F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ9
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2463 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZA
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ8
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L184F/A187F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZD
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant L184F/A187F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ1
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-13 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
1T7D
 
 | | Crystal structure of Escherichia coli type I signal peptidase in complex with a lipopeptide inhibitor | | Descriptor: | 10-METHYLUNDECANOIC ACID, ARYLOMYCIN A2, SIGNAL PEPTIDASE I | | Authors: | Paetzel, M, Goodall, J.J, Kania, M, Dalbey, R.E, Page, M.G.P. | | Deposit date: | 2004-05-09 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystallographic and Biophysical Analysis of a Bacterial Signal Peptidase in Complex with a Lipopeptide Based Inhibitor.
J.Biol.Chem., 279, 2004
|
|
2LMK
 
 | | Solution Structure of Mouse Pheromone ESP1 | | Descriptor: | Exocrine gland-secreting peptide 1 | | Authors: | Yoshinaga, S, Sato, T, Hirakane, M, Esaki, K, Hamaguchi, T, Haga-Yamanaka, S, Tsunoda, M, Kimoto, H, Shimada, I, Touhara, K, Terasawa, H. | | Deposit date: | 2011-12-06 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure of the Mouse Sex Peptide Pheromone ESP1 Reveals a Molecular Basis for Specific Binding to the Class-C G-Protein-Coupled Vomeronasal Receptor
J.Biol.Chem., 2013
|
|
6FZ7
 
 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L184F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6TF7
 
 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | 4-fluoranyl-~{N}-[[(2~{S},3~{R},4~{R},5~{R},6~{R})-6-(hydroxymethyl)-3,5-bis(oxidanyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxan-2-yl]methyl]naphthalene-1-carboxamide, CHLORIDE ION, Galectin-3, ... | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
6TF6
 
 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | CHLORIDE ION, Galectin-3, ~{N}-[[(2~{S},3~{S},4~{R},5~{S},6~{R})-4-[[5,6-bis(fluoranyl)-2-oxidanylidene-chromen-3-yl]methoxy]-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]methyl]-4-fluoranyl-naphthalene-1-carboxamide | | Authors: | Nilsson, U.J, Zetterberg, F, Hakansson, M, Logan, D.T. | | Deposit date: | 2019-11-13 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 3-Substituted 1-Naphthamidomethyl-C-galactosyls Interact with Two Unique Sub-sites for High-Affinity and High-Selectivity Inhibition of Galectin-3.
Molecules, 24, 2019
|
|
2FAW
 
 | | crystal structure of papaya glutaminyl cyclase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, ZINC ION, ... | | Authors: | Wintjens, R, Belrhali, H, Clantin, B, Azarkan, M, Bompard, C, Baeyens-Volant, D, Looze, Y, Villeret, V. | | Deposit date: | 2005-12-08 | | Release date: | 2006-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of papaya glutaminyl cyclase, an archetype for plant and bacterial glutaminyl cyclases.
J.Mol.Biol., 357, 2006
|
|
7F1M
 
 | | Marburg virus nucleoprotein-RNA complex | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Fujita, F.Y, Sugita, Y, Takamatsu, Y, Houri, K, Muramoto, Y, Nakano, M, Tsunoda, Y, Igarashi, M, Becker, S, Noda, T. | | Deposit date: | 2021-06-09 | | Release date: | 2022-03-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insight into Marburg virus nucleoprotein-RNA complex formation.
Nat Commun, 13, 2022
|
|
1WP1
 
 | | Crystal structure of the drug-discharge outer membrane protein, OprM | | Descriptor: | Outer membrane protein oprM | | Authors: | Akama, H, Kanemaki, M, Yoshimura, M, Tsukihara, T, Kashiwagi, T, Narita, S, Nakagawa, A, Nakae, T. | | Deposit date: | 2004-08-28 | | Release date: | 2004-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: dual modes of membrane anchoring and occluded cavity end
J.Biol.Chem., 279, 2004
|
|
4HD6
 
 | | Crystal Structure of Tyrosinase from Bacillus megaterium V218F mutant soaked in CuSO4 | | Descriptor: | COPPER (II) ION, Tyrosinase | | Authors: | Goldfeder, M, Kanteev, M, Adir, N, Fishman, A. | | Deposit date: | 2012-10-02 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Influencing the monophenolase/diphenolase activity ratio in tyrosinase.
Biochim.Biophys.Acta, 1834, 2013
|
|
