3LHK
 
 | |
3LSO
 
 | |
6DWE
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate- and BRD0059-bound form | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-26 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
3LUP
 
 | | Crystal structure of fatty acid binding DegV family protein SAG1342 from Streptococcus agalactiae | | Descriptor: | 9-OCTADECENOIC ACID, DegV family protein, GLYCEROL | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-02 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of fatty acid binding DegV family protein SAG1342 from Streptococcus agalactiae
To be Published
|
|
3LVU
 
 | | Crystal structure of ABC transporter, periplasmic substrate-binding protein SPO2066 from Silicibacter pomeroyi | | Descriptor: | 1,2-ETHANEDIOL, 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, ABC transporter, ... | | Authors: | Chang, C, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of ABC transporter, periplasmic substrate-binding protein SPO2066 from Silicibacter pomeroyi
To be Published
|
|
1N1F
 
 | | Crystal Structure of Human Interleukin-19 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, interleukin-19 | | Authors: | Chang, C, Magracheva, E, Kozlov, S, Fong, S, Tobin, G, Kotenko, S, Wlodawer, A, Zdanov, A. | | Deposit date: | 2002-10-17 | | Release date: | 2003-02-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of interleukin-19 defines a new subfamily of helical cytokines
J.Biol.Chem., 278, 2003
|
|
3JU3
 
 | | Crystal structure of alpha chain of probable 2-oxoacid ferredoxin oxidoreductase from Thermoplasma acidophilum | | Descriptor: | 1,2-ETHANEDIOL, Probable 2-oxoacid ferredoxin oxidoreductase, alpha chain | | Authors: | Chang, C, Marshall, N, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of alpha chain of probable 2-oxoacid ferredoxin oxidoreductase from Thermoplasma acidophilum
To be Published
|
|
3CWF
 
 | | Crystal structure of PAS domain of two-component sensor histidine kinase | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alkaline phosphatase synthesis sensor protein phoR | | Authors: | Chang, C, Tesar, C, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-21 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Extracytoplasmic PAS-like domains are common in signal transduction proteins.
J.Bacteriol., 192, 2010
|
|
3K67
 
 | | Crystal structure of protein af1124 from archaeoglobus fulgidus | | Descriptor: | PHOSPHATE ION, putative dehydratase AF1124 | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal Structure of Protein Af1124 from Archaeoglobus Fulgidus
To be Published
|
|
6E9P
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - open form with BRD0059 bound | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-08-01 | | Release date: | 2018-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.569 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
3K6R
 
 | | CRYSTAL STRUCTURE OF putative transferase PH0793 FROM PYROCOCCUS HORIKOSHII | | Descriptor: | putative transferase PH0793 | | Authors: | Chang, C, Skarina, T, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-09 | | Release date: | 2009-10-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Protein Ph0793 from Pyrococcus Horikoshii
To be Published
|
|
3KAO
 
 | | Crystal structure of tagatose 1,6-diphosphate aldolase from Staphylococcus aureus | | Descriptor: | GLYCEROL, SULFATE ION, Tagatose 1,6-diphosphate aldolase, ... | | Authors: | Chang, C, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-19 | | Release date: | 2009-10-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of tagatose 1,6-diphosphate aldolase from Staphylococcus aureus
To be Published
|
|
3K6C
 
 | | Crystal structure of protein ne0167 from nitrosomonas europaea | | Descriptor: | Uncharacterized protein NE0167 | | Authors: | Chang, C, Evdokimova, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Protein Ne0167 from Nitrosomonas Europaea
To be Published
|
|
3K6A
 
 | |
6DIN
 
 | | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase | | Descriptor: | SULFATE ION, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Chang, C, Jedrzejczak, R, Chhor, G, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-23 | | Release date: | 2018-05-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolutionstructure of apo dTDP-4-dehydrorhamnose 3,5-epimerase
To Be Published
|
|
3BEE
 
 | | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus | | Descriptor: | 1,2-ETHANEDIOL, Putative YfrE protein | | Authors: | Chang, C, Hatzos, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-11-16 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative YfrE protein from Vibrio parahaemolyticus.
To be Published
|
|
3KWP
 
 | | Crystal structure of putative methyltransferase from Lactobacillus brevis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Predicted methyltransferase | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-12-01 | | Release date: | 2009-12-15 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of putative methyltransferase from Lactobacillus brevis
To be Published
|
|
3MT1
 
 | | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti | | Descriptor: | Putative carboxynorspermidine decarboxylase protein, SULFATE ION | | Authors: | Chang, C, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-04-29 | | Release date: | 2010-06-30 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of putative carboxynorspermidine decarboxylase protein from Sinorhizobium meliloti
To be Published
|
|
3M0Z
 
 | | Crystal structure of putative aldolase from Klebsiella pneumoniae. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, SULFATE ION, ... | | Authors: | Chang, C, Rakowski, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-03-03 | | Release date: | 2010-03-31 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of putative aldolase from Klebsiella pneumoniae.
To be Published
|
|
3DNP
 
 | | Crystal structure of Stress response protein yhaX from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Stress response protein yhaX | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-07-29 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Stress response protein yhaX from Bacillus subtilis
To be Published
|
|
3MGL
 
 | |
5JBR
 
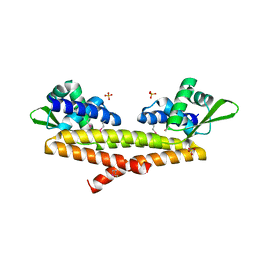 | | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae | | Descriptor: | SULFATE ION, Uncharacterized protein Bcav_2135 | | Authors: | Chang, C, Cuff, M, Hatzos-Skintges, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-04-13 | | Release date: | 2016-04-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of uncharacterized protein Bcav_2135 from Beutenbergia cavernae
To Be Published
|
|
5JH8
 
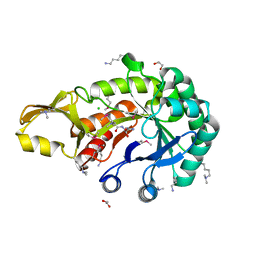 | | Crystal structure of chitinase from Chromobacterium violaceum ATCC 12472 | | Descriptor: | (2S)-2-(dimethylamino)-4-(methylselanyl)butanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Chang, C, Michalska, K, Tesar, C, Clancy, S, Joachimiak, A. | | Deposit date: | 2016-04-20 | | Release date: | 2016-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.018 Å) | | Cite: | Crystal structure of chitinase from Chromobacterium violaceum ATCC 12472
To Be Published
|
|
5K9X
 
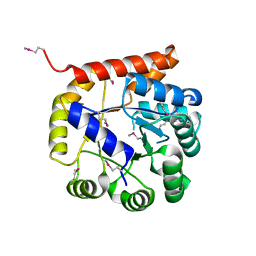 | | Crystal structure of Tryptophan synthase alpha chain from Legionella pneumophila subsp. pneumophila | | Descriptor: | Tryptophan synthase alpha chain | | Authors: | Chang, C, Hatzos-Skintges, C, Endres, M, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-01 | | Release date: | 2016-06-15 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Crystal structure of Tryptophan synthase alpha chain from Legionella pneumophila subsp. pneumophila
To Be Published
|
|
3MXQ
 
 | |
