6QUB
 
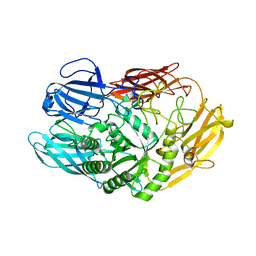 | | Truncated beta-galactosidase III from Bifidobacterium bifidum in complex with galactose | | Descriptor: | Beta-galactosidase, CALCIUM ION, beta-D-galactopyranose | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
6QUC
 
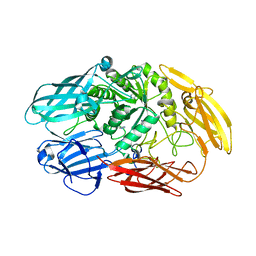 | | Truncated beta-galactosidase III from Bifidobacterium bifidum | | Descriptor: | Beta-galactosidase, CALCIUM ION, IMIDAZOLE | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
4AXW
 
 | | CRYSTAL STRUCTURE OF MOUSE CADHERIN-23 EC1-2 AND PROTOCADHERIN-15 EC1- 2, FORM I 2.2A. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CADHERIN-23, CALCIUM ION, ... | | Authors: | Sotomayor, M, Weihofen, W, Gaudet, R, Corey, D.P. | | Deposit date: | 2012-06-14 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure of a Force-Conveying Cadherin Bond Essential for Inner-Ear Mechanotransduction
Nature, 492, 2012
|
|
4B3O
 
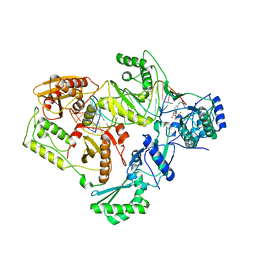 | | Structures of HIV-1 RT and RNA-DNA Complex Reveal a Unique RT Conformation and Substrate Interface | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, 5'-D(*CP*GP*TP*AP*TP*GP*CP*CP*TP*AP*TP*AP*GP*TP *TP*AP*TP*TP*GP*TP*GP*GP*CP*C)-3', 5'-R(*AP*UP*GP*AP*3DRP*GP*GP*CP*CP*AP*CP*AP*AP*UP*AP *AP*CP*UP*AP*UP*AP*GP*GP*CP*AP*UP*A)-3', ... | | Authors: | Lapkouski, M, Tian, L, Miller, J.T, Le Grice, S.F.J, Yang, W. | | Deposit date: | 2012-07-25 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Complexes of HIV-1 RT, Nnrti and RNA/DNA Hybrid Reveal a Structure Compatible with RNA Degradation
Nat.Struct.Mol.Biol., 20, 2013
|
|
4B41
 
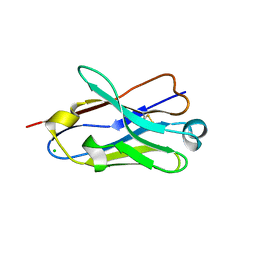 | | Crystal structure of an amyloid-beta binding single chain antibody G7 | | Descriptor: | ANTIBODY G7, CHLORIDE ION, GLYCEROL | | Authors: | Beringer, D.X, Dorresteijn, B, Rutten, L, Wienk, H, el Khattabi, M, Kroon-Batenburg, L.M.J, Verrips, C.T. | | Deposit date: | 2012-07-27 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.191 Å) | | Cite: | Crystal Structure of an Amyloid-Beta Binding Single Chain Antibody G7
To be Published
|
|
4B65
 
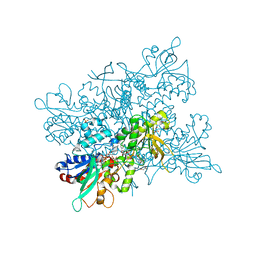 | | A. fumigatus ornithine hydroxylase (SidA), reduced state bound to NADP(H) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, L-ORNITHINE N5 MONOOXYGENASE, ... | | Authors: | Franceschini, S, Fedkenheuer, M, Vogelaar, N.J, Robinson, H.H, Sobrado, P, Mattevi, A. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Insight Into the Mechanism of Oxygen Activation and Substrate Selectivity of Flavin-Dependent N-Hydroxylating Monooxygenases.
Biochemistry, 51, 2012
|
|
6QW2
 
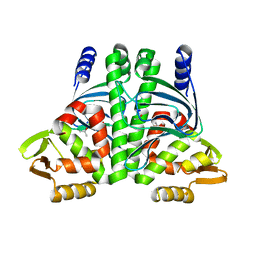 | | The Transcriptional Regulator PrfA-A218G mutant from Listeria Monocytogenes | | Descriptor: | ISOPROPYL ALCOHOL, Listeriolysin positive regulatory factor A, SODIUM ION | | Authors: | Hall, M, Grundstrom, C, Hansen, S, Brannstrom, K, Johansson, J, Sauer-Eriksson, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Novel Growth-Based Selection Strategy Identifies New Constitutively Active Variants of the Major Virulence Regulator PrfA in Listeria monocytogenes.
J.Bacteriol., 202, 2020
|
|
4B3M
 
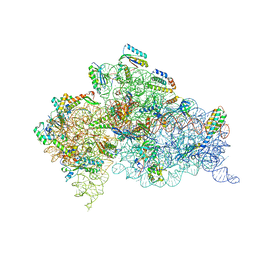 | | Crystal structure of the 30S ribosome in complex with compound 1 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4,6-O-benzylidene-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-25 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4B66
 
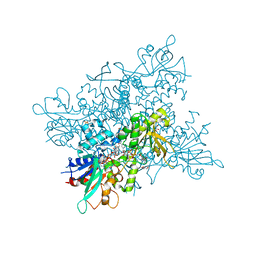 | | A. fumigatus ornithine hydroxylase (SidA), reduced state bound to NADP and Arg | | Descriptor: | ARGININE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Franceschini, S, Fedkenheuer, M, Vogelaar, N.J, Robinson, H.H, Sobrado, P, Mattevi, A. | | Deposit date: | 2012-08-09 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Insight Into the Mechanism of Oxygen Activation and Substrate Selectivity of Flavin-Dependent N-Hydroxylating Monooxygenases.
Biochemistry, 51, 2012
|
|
4B8W
 
 | | Crystal structure of human GDP-L-fucose synthase with bound NADP and GDP, tetragonal crystal form | | Descriptor: | GDP-L-FUCOSE SYNTHASE, GUANOSINE-5'-DIPHOSPHATE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Vollmar, M, Krojer, T, Shafqat, N, Rojkova, A, Bunkoczi, G, Pike, A, Chaikuat, A, Carpenter, E, Yue, W.W, Kavanagh, K, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2012-08-31 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of Human Gdp-L-Fucose Synthase with Bound Nadp and Gdp, Tetragonal Crystal Form
To be Published
|
|
4URA
 
 | | Crystal structure of human JMJD2A in complex with compound 14a | | Descriptor: | 1,2-ETHANEDIOL, 2-(2H-1,2,3-triazol-4-yl)pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Krojer, T, England, K.S, Vollmar, M, Crawley, L, Williams, E, Riesebos, E, Szykowska, A, Burgess-Brown, N, Oppermann, U, Brennan, P.E, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2014-06-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Optimisation of a triazolopyridine based histone demethylase inhibitor yields a potent and selective KDM2A (FBXL11) inhibitor.
Medchemcomm, 5, 2014
|
|
4BAY
 
 | | Phosphomimetic mutant of LSD1-8a splicing variant in complex with CoREST | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1 | | Authors: | Toffolo, E, Paganini, L, Rusconi, F, Tortorici, M, Pilotto, S, Verpelli, C, Tedeschi, G, Maffioli, E, Sala, C, Mattevi, A, Battaglioli, E. | | Deposit date: | 2012-09-17 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phosphorylation of Neuronal Lysine-Specific Demethylase 1Lsd1/Kdm1A Impairs Transcriptional Repression by Regulating Interaction with Corest and Histone Deacetylases Hdac1/2.
J.Neurochem., 128, 2014
|
|
4BF2
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | ACETATE ION, GLYCEROL, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, ... | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-03-13 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4BCB
 
 | | PROLYL OLIGOPEPTIDASE FROM PORCINE BRAIN WITH A COVALENTLY BOUND P2- substituted N-acyl-prolylpyrrolidine inhibitor | | Descriptor: | (5R,6R,8S)-8-(3-{[AMINO(IMINO)METHYL]AMINO}PHENYL)-5-CYCLOHEXYL-6-HYDROXY-3-OXO-1-PHENYL-2,7-DIOXA-4-AZA-6-PHOSPHANONAN-9-OIC ACID 6-OXIDE, GLYCEROL, PROLYL ENDOPEPTIDASE, ... | | Authors: | VanDerVeken, P, Fulop, V, Rea, D, Gerard, M, VanElzen, R, Joossens, J, Cheng, J.D, Baekelandt, V, DeMeester, I, Lambeir, A.M, Augustyns, K. | | Deposit date: | 2012-10-01 | | Release date: | 2013-03-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P2-Substituted N-Acylprolylpyrrolidine Inhibitors of Prolyl Oligopeptidase: Biochemical Evaluation, Binding Mode Determination, and Assessment in a Cellular Model of Synucleinopathy.
J.Med.Chem., 55, 2012
|
|
4BDB
 
 | | Fragment-based screening identifies a new area for inhibitor binding to checkpoint kinase 2 (CHK2) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-C-methyl-N-oxidanyl-carbonimidoyl]benzene-1,3-diol, NITRATE ION, ... | | Authors: | Silva-Santisteban, M.C, Westwood, I.M, Boxall, K, Brown, N, Peacock, S, McAndrew, C, Barrie, E, Richards, M, Mirza, A, Oliver, A.W, Burke, R, Hoelder, S, Jones, K, Aherne, G.W, Blagg, J, Collins, I, Garrett, M.D, van Montfort, R.L.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screening Maps Inhibitor Interactions in the ATP-Binding Site of Checkpoint Kinase 2.
Plos One, 8, 2013
|
|
4BID
 
 | | Crystal Structures of Ask1-inhibitor Complexes | | Descriptor: | 5-[(1R)-2-amino-1-phenylethoxy]-2-(furan-3-yl)-3-{1H-pyrrolo[2,3-b]pyridin-3-yl}pyridine, ACETATE ION, MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5 | | Authors: | Singh, O, Shillings, A, Craggs, P, Wall, I, Rowland, P, Skarzynski, T, Hobbs, C.I, Hardwick, P, Tanner, R, Blunt, M, Witty, D.R, Smith, K.J. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structures of Ask1-Inhibtor Complexes Provide a Platform for Structure Based Drug Design.
Protein Sci., 22, 2013
|
|
4AS4
 
 | | Structure of human inositol monophosphatase 1 | | Descriptor: | GLYCEROL, INOSITOL MONOPHOSPHATASE 1, MAGNESIUM ION, ... | | Authors: | Singh, N, Knight, M, Halliday, A.C, Lack, N.A, Lowe, E.D, Churchill, G.C. | | Deposit date: | 2012-04-27 | | Release date: | 2012-10-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Cloning, Expression, Purification, Crystallization and X-Ray Analysis of Inositol Monophosphatase from Mus Musculus and Homo Sapiens.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
4BE6
 
 | |
4BFL
 
 | |
4AYD
 
 | | Structure of a complex between CCPs 6 and 7 of Human Complement Factor H and Neisseria meningitidis FHbp Variant 1 R106A mutant | | Descriptor: | 1,2-ETHANEDIOL, COMPLEMENT FACTOR H, FACTOR H BINDING PROTEIN | | Authors: | Johnson, S, Tan, L, van der Veen, S, Caesar, J, Goicoechea De Jorge, E, Everett, R.J, Bai, X, Exley, R.M, Ward, P.N, Ruivo, N, Trivedi, K, Cumber, E, Jones, R, Newham, L, Staunton, D, Borrow, R, Pickering, M, Lea, S.M, Tang, C.M. | | Deposit date: | 2012-06-20 | | Release date: | 2012-11-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and Evaluation of Meningococcal Vaccines Through Structure-Based Modification of Host and Pathogen Molecules
Plos Pathog., 8, 2012
|
|
6QX7
 
 | |
4B1B
 
 | | Crystal structure of Plasmodium falciparum oxidised Thioredoxin Reductase at 2.9 angstrom | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THIOREDOXIN REDUCTASE | | Authors: | Boumis, G, Giardina, G, Dimastrogiovanni, D, Angelucci, F, Saccoccia, F, Brunori, M, Bellelli, A, Miele, A.E. | | Deposit date: | 2012-07-09 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Plasmodium Falciparum Thioredoxin Reductase, a Validated Drug Target.
Biochem.Biophys.Res.Commun., 425, 2012
|
|
4ASE
 
 | | CRYSTAL STRUCTURE OF VEGFR2 (JUXTAMEMBRANE AND KINASE DOMAINS) IN COMPLEX WITH TIVOZANIB (AV-951) | | Descriptor: | TIVOZANIB, VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 2 | | Authors: | McTigue, M, Deng, Y, Ryan, K, Brooun, A, Diehl, W, Stewart, A. | | Deposit date: | 2012-04-30 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular Conformations, Interactions, and Properties Associated with Drug Efficiency and Clinical Performance Among Vegfr Tk Inhibitors.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B3R
 
 | | Crystal structure of the 30S ribosome in complex with compound 30 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-2-deoxy-4,6-O-[(1R)-3-phenylpropylidene]-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
4AVX
 
 | | Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate (Hgs-Hrs) bound to an IP2 compound at 1.68 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, HEPATOCYTE GROWTH FACTOR-REGULATED TYROSINE KINASE SUBSTRATE, PHOSPHORIC ACID MONO-(2,3,4,6-TETRAHYDROXY-5-PHOSPHONOOXY-CYCLOHEXYL) ESTER, ... | | Authors: | Williams, E, Canning, P, Shrestha, L, Krojer, T, Vollmar, M, Slowey, A, Conway, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bullock, A. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of the Tandem Vhs and Fyve Domains of Hepatocyte Growth Factor-Regulated Tyrosine Kinase Substrate (Hgs-Hrs) Bound to an Ip2 Compound at 1.68 A Resolution
To be Published
|
|
