1XZA
 
 | |
5Z1R
 
 | | Crystal Structure Analysis of the BRD4 | | Descriptor: | 1,2-ETHANEDIOL, 5-bromo-N-(2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)-2-methoxybenzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | Deposit date: | 2017-12-28 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
6ORK
 
 | | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes | | Descriptor: | Sel1 repeat protein | | Authors: | Chang, C, Tesar, C, Endres, M, Babnigg, G, Hassan, H, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2019-04-30 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of Sel1 repeat protein from Oxalobacter formigenes
To Be Published
|
|
6EML
 
 | | Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S11-A, ... | | Authors: | Heuer, A, Thomson, E, Schmidt, C, Berninghausen, O, Becker, T, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of a late pre-40S ribosomal subunit fromSaccharomyces cerevisiae.
Elife, 6, 2017
|
|
8J2P
 
 | | Crystal structure of PML B-box2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-15 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
8J25
 
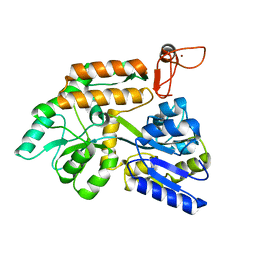 | | Crystal structure of PML B-box2 mutant | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Protein PML, ZINC ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Zhou, C, Zang, N, Zhang, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-09-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of PML-RARA Oncoprotein Targeting by Arsenic Unravels a Cysteine Rheostat Controlling PML Body Assembly and Function.
Cancer Discov, 13, 2023
|
|
6EBK
 
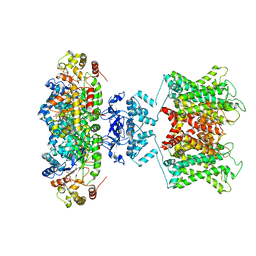 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6EPF
 
 | | Ground state 26S proteasome (GS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EDF
 
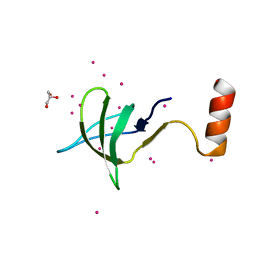 | | Fragment of a tyrosine-protein kinase | | Descriptor: | FYN, GLYCEROL, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Huang, H, Sochirca, I, Liu, K, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Sidhu, S.S, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-08-09 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Fragment of a tyrosine-protein kinase
To be Published
|
|
6EQC
 
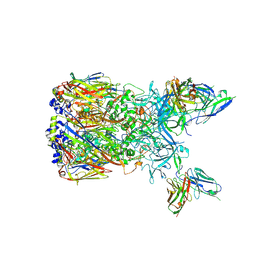 | | Cryo-EM reconstruction of a complex of a binding protein and human adenovirus C5 hexon | | Descriptor: | Hexon protein, scFv of 9C12 antibody | | Authors: | Schmid, M, Ernst, P, Honegger, A, Suomalainen, M, Zimmermann, M, Braun, L, Stauffer, S, Thom, C, Dreier, B, Eibauer, M, Kipar, A, Vogel, V, Greber, U.F, Medalia, O, Plueckthun, A. | | Deposit date: | 2017-10-12 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Adenoviral vector with shield and adapter increases tumor specificity and escapes liver and immune control.
Nat Commun, 9, 2018
|
|
6EMN
 
 | | HtxB from Pseudomonas stutzeri in complex with phosphite to 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Probable phosphite transport system-binding protein HtxB, ... | | Authors: | Bisson, C, Robertson, A.J, Hitchcock, A, Adams, N.B. | | Deposit date: | 2017-10-03 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphite binding by the HtxB periplasmic binding protein depends on the protonation state of the ligand.
Sci Rep, 9, 2019
|
|
6EQI
 
 | | Structure of PINK1 bound to ubiquitin | | Descriptor: | GLYCEROL, Nb696, Serine/threonine-protein kinase PINK1, ... | | Authors: | Schubert, A.F, Gladkova, C, Pardon, E, Wagstaff, J.L, Freund, S.M.V, Steyaert, J, Maslen, S, Komander, D. | | Deposit date: | 2017-10-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of PINK1 in complex with its substrate ubiquitin.
Nature, 552, 2017
|
|
6EM4
 
 | | State B architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | Authors: | Kater, L, Cheng, J, Barrio-Garcia, C, Hurt, E, Beckmann, R. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Visualizing the Assembly Pathway of Nucleolar Pre-60S Ribosomes.
Cell, 171, 2017
|
|
6EPE
 
 | | Substrate processing state 26S proteasome (SPS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
5UE3
 
 | | proMMP-9desFnII | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Matrix metalloproteinase-9, ... | | Authors: | Alexander, R.S, Spurlino, J, Milligan, C. | | Deposit date: | 2016-12-29 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Discovery of a highly selective chemical inhibitor of matrix metalloproteinase-9 (MMP-9) that allosterically inhibits zymogen activation.
J. Biol. Chem., 292, 2017
|
|
6PB0
 
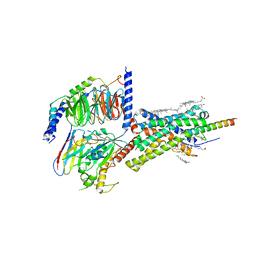 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
6ELY
 
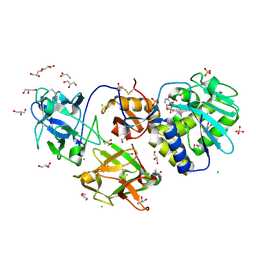 | | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.84 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-N-Furfurylcytosine, ... | | Authors: | Ahmad, M.S, Rasheed, S, Falke, S, Khaliq, B, Perbandt, M, Choudhary, M.I, Markiewicz, W.T, Barciszewski, J, Betzel, C. | | Deposit date: | 2017-09-30 | | Release date: | 2018-05-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal Structure of Mistletoe Lectin I (ML-I) from Viscum album in Complex with 4-N-Furfurylcytosine at 2.85 angstrom Resolution.
Med Chem, 14, 2018
|
|
6EN4
 
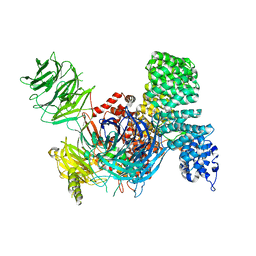 | | SF3b core in complex with a splicing modulator | | Descriptor: | PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, Splicing factor 3B subunit 3, ... | | Authors: | Cretu, C, Pena, V. | | Deposit date: | 2017-10-04 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural Basis of Splicing Modulation by Antitumor Macrolide Compounds.
Mol. Cell, 70, 2018
|
|
6EBL
 
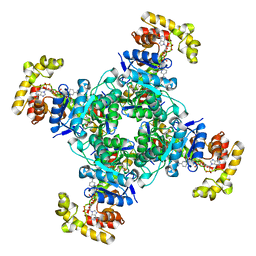 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs, cytosolic domain | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | Authors: | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | Deposit date: | 2018-08-06 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
6EQS
 
 | | Human Sirt5 in complex with stalled peptidylimidate intermediate of inhibitory compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-BUTANEDIOL, 3-[[(~{Z})-~{C}-[(2~{R},3~{R},4~{S},5~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-3,4-bis(oxidanyl)oxolan-2-yl]sulfanyl-~{N}-[(5~{S})-6-[[(2~{S})-3-(1~{H}-indol-3-yl)-1-oxidanylidene-1-(propan-2-ylamino)propan-2-yl]amino]-6-oxidanylidene-5-(phenylmethoxycarbonylamino)hexyl]carbonimidoyl]amino]propanoic acid, ... | | Authors: | Pannek, M, Steegborn, C. | | Deposit date: | 2017-10-15 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Mechanism-Based Inhibitors of the Human Sirtuin 5 Deacylase: Structure-Activity Relationship, Biostructural, and Kinetic Insight.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6ENX
 
 | | Zebrafish Sirt5 in complex with stalled bicyclic intermediate of inhibitory compound 10 | | Descriptor: | 4-[(2~{R},3~{a}~{R},5~{R},6~{R},6~{a}~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2-[[(5~{S})-6-[[(2~{S})-3-(1~{H}-indol-3-yl)-1-oxidanylidene-1-(propan-2-ylamino)propan-2-yl]amino]-6-oxidanylidene-5-(phenylmethoxycarbonylamino)hexyl]amino]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]butanoic acid, DIMETHYL SULFOXIDE, NAD-dependent protein deacylase sirtuin-5, ... | | Authors: | Pannek, M, Steegborn, C. | | Deposit date: | 2017-10-06 | | Release date: | 2017-11-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism-Based Inhibitors of the Human Sirtuin 5 Deacylase: Structure-Activity Relationship, Biostructural, and Kinetic Insight.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6E4J
 
 | | Solution NMR Structure of Protein PF2048.1 | | Descriptor: | Uncharacterized protein PF2048.1 | | Authors: | Daigham, N.S, Liu, G, Swapna, G.V.T, Cole, C, Valafar, H, Montelione, G.T. | | Deposit date: | 2018-07-17 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | REDCRAFT: A Computational Platform Using Residual Dipolar Coupling NMR Data for Determining Structures of Perdeuterated Proteins Without NOEs
To Be Published
|
|
6EHZ
 
 | |
6E98
 
 | |
6E9M
 
 | |
