8IVA
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs XMA01 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV8
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 3E2 and 1C4 (local refinement) | | Descriptor: | Spike protein S1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, heavy chain of 1C4, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8IV4
 
 | | Cryo-EM structure of SARS-CoV-2 spike protein in complex with double nAbs 8H12 and 3E2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, heavy chain of 3E2, ... | | Authors: | Sun, H, Jiang, Y, Zheng, Q, Li, S, Xia, N. | | Deposit date: | 2023-03-26 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Two antibodies show broad, synergistic neutralization against SARS-CoV-2 variants by inducing conformational change within the RBD.
Protein Cell, 15, 2024
|
|
8QCX
 
 | |
8QCS
 
 | |
8QCY
 
 | |
8QD0
 
 | |
8QCT
 
 | |
3CTJ
 
 | |
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | Authors: | Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
7KN4
 
 | |
4R89
 
 | | Crystal structure of paFAN1 - 5' flap DNA complex with Manganase | | Descriptor: | DNA (5'-D(P*AP*CP*CP*AP*GP*AP*CP*AP*CP*AP*CP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*C)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Cho, Y, Gwon, G.H, Kim, Y.R. | | Deposit date: | 2014-08-30 | | Release date: | 2014-10-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.002 Å) | | Cite: | Crystal structure of a Fanconi anemia-associated nuclease homolog bound to 5' flap DNA: basis of interstrand cross-link repair by FAN1
Genes Dev., 28, 2014
|
|
8D1G
 
 | | hBest2 Ca2+-bound open state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.07 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1I
 
 | | hBest1 1uM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.82 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1O
 
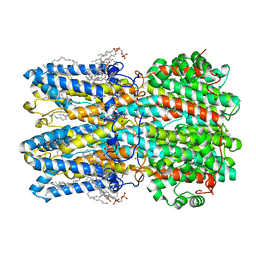 | | hBest1_345 Ca2+-bound open state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1K
 
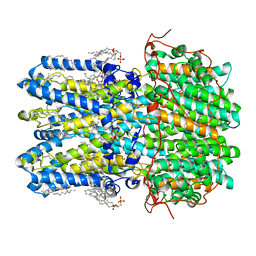 | | hBest1 Ca2+-bound partially open neck state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1N
 
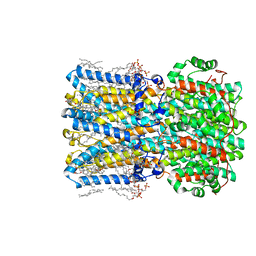 | | bBest2_345 Ca2+-bound open state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1F
 
 | | hBest2 5mM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.82 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1E
 
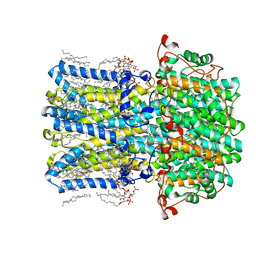 | | hBest2 1uM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.78 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1L
 
 | | hBest1 Ca2+-bound partially open aperture state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1J
 
 | | hBest1 5mM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.05 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
8D1M
 
 | |
8D1H
 
 | | hBest2 Ca2+-unbound closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, 2-[2-[(1~{S},2~{S},4~{S},5'~{R},6~{R},7~{S},8~{R},9~{S},12~{S},13~{R},16~{S})-5',7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icos-18-ene-6,2'-oxane]-16-yl]oxyethyl]propane-1,3-diol, Bestrophin-2, ... | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
6ZOV
 
 | | ENTEROPEPTIDASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-carbamimidamidobenzoic acid, Enteropeptidase, ... | | Authors: | Cummings, M.D. | | Deposit date: | 2020-07-07 | | Release date: | 2020-10-21 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Targeting Enteropeptidase with Reversible Covalent Inhibitors To Achieve Metabolic Benefits.
J.Pharmacol.Exp.Ther., 375, 2020
|
|
7C01
 
 | | Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 heavy chain, CB6 light chain, ... | | Authors: | Shi, R, Qi, J, Wang, Q, Gao, F.G, Yan, J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2.
Nature, 584, 2020
|
|
