3V9R
 
 | | Crystal structure of Saccharomyces cerevisiae MHF complex | | Descriptor: | SULFATE ION, Uncharacterized protein YDL160C-A, Uncharacterized protein YOL086W-A | | Authors: | Yang, H, Zhang, T, Zhong, C, Li, H, Zhou, J, Ding, J. | | Deposit date: | 2011-12-28 | | Release date: | 2012-02-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Saccharomyces Cerevisiae MHF Complex Structurally Resembles the Histones (H3-H4)(2) Heterotetramer and Functions as a Heterotetramer
Structure, 20, 2012
|
|
4Z62
 
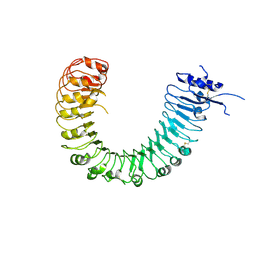 | | The plant peptide hormone free receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Phytosulfokine receptor 1 | | Authors: | Chai, J, Wang, J, Han, Z. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
4Z5W
 
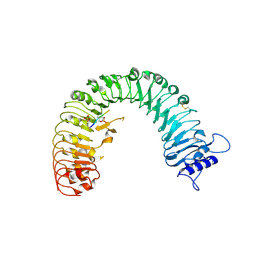 | | The plant peptide hormone receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phytosulfokine, ... | | Authors: | Chai, J, Wang, J, Han, Z. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
4Z61
 
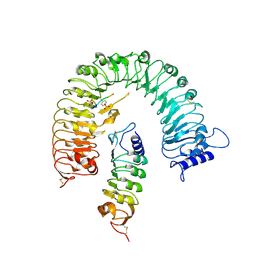 | | The plant peptide hormone receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PTR-ILE-PTR-THR-GLN, Phytosulfokine receptor 1, ... | | Authors: | Chai, J, Wang, J. | | Deposit date: | 2015-04-03 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Allosteric receptor activation by the plant peptide hormone phytosulfokine
Nature, 525, 2015
|
|
5WTD
 
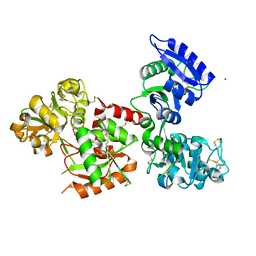 | | Structure of human serum transferrin bound ruthenium at N-lobe | | Descriptor: | FE (III) ION, MALONATE ION, RUTHENIUM ION, ... | | Authors: | Sun, H, Wang, M, Lai, T.P, Zhang, H, Hao, Q. | | Deposit date: | 2016-12-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Binding of ruthenium and osmium at non‐iron sites of transferrin accounts for their iron-independent cellular uptake.
J.Inorg.Biochem., 234, 2022
|
|
4D3O
 
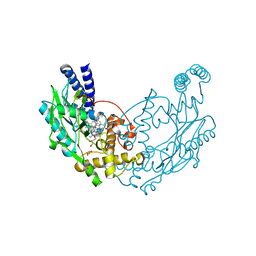 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6-(3-(2-(1H-Pyrrolo(2,3-b)pyridin-6-yl)ethyl)-5-(aminomethyl) phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 6-(2-{3-(aminomethyl)-5-[2-(1H-pyrrolo[2,3-b]pyridin-6-yl)ethyl]phenyl}ethyl)-4-methylpyridin-2-amine, CHLORIDE ION, GLYCEROL, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-23 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
4D3V
 
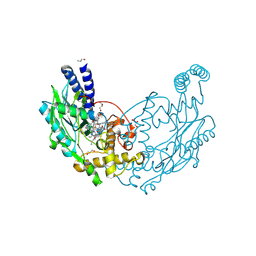 | |
4D3M
 
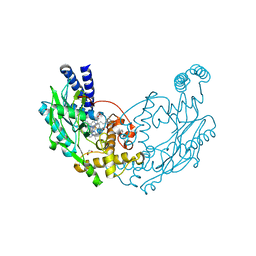 | |
4D3J
 
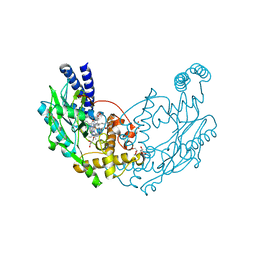 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6,6'-(2,2'-(5-amino-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 6,6'-[(5-aminobenzene-1,3-diyl)diethane-2,1-diyl]bis(4-methylpyridin-2-amine), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
4D3K
 
 | | Structure of Bacillus subtilis nitric oxide synthase in complex with 6,6'-((5-(3-aminopropyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 6,6'-{[5-(3-aminopropyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.017 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
4D3N
 
 | |
4D3U
 
 | |
4D3I
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6,6'-((5-(aminomethyl)-1,3-phenylene)bis(ethane-2,1-diyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 6,6'-{[5-(aminomethyl)benzene-1,3-diyl]diethane-2,1-diyl}bis(4-methylpyridin-2-amine), GLYCEROL, N-PROPANOL, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Based Design of Bacterial Nitric Oxide Synthase Inhibitors.
J.Med.Chem., 58, 2015
|
|
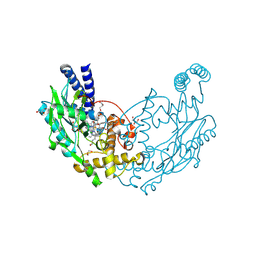
4D3T
 
 | |
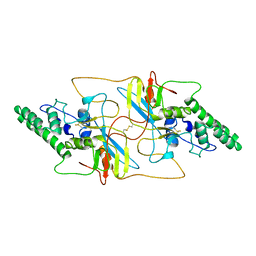
8I8A
 
 | |
8I8C
 
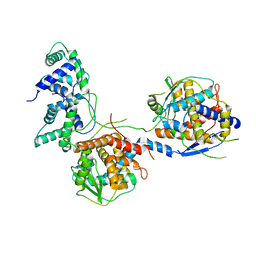 | |
8I8B
 
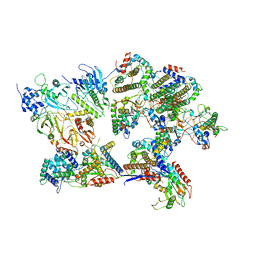 | | Outer shell and inner layer structures of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) | | Descriptor: | 38K, AcOrf-109 peptide, Early 49 Daa protein, ... | | Authors: | Jia, X, Gao, Y, Zhang, Q. | | Deposit date: | 2023-02-03 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Architecture of the baculovirus nucleocapsid revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
5I78
 
 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
5I79
 
 | | Crystal structure of a beta-1,4-endoglucanase mutant from Aspergillus niger in complex with sugar | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Endo-beta-1, ... | | Authors: | Liu, W.D, Yan, J.J, Li, Y.J, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
6LNA
 
 | | YdiU complex with AMPNPP and Mn2+ | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Li, B, Yang, Y, Ma, Y. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | The YdiU Domain Modulates Bacterial Stress Signaling through Mn 2+ -Dependent UMPylation.
Cell Rep, 32, 2020
|
|
3RFZ
 
 | | Crystal structure of the FimD usher bound to its cognate FimC:FimH substrate | | Descriptor: | Chaperone protein fimC, Outer membrane usher protein, type 1 fimbrial synthesis, ... | | Authors: | Phan, G, Remaut, H, Lebedev, A, Geibel, S, Waksman, G. | | Deposit date: | 2011-04-07 | | Release date: | 2011-06-01 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the FimD usher bound to its cognate FimC-FimH substrate.
Nature, 474, 2011
|
|
5I77
 
 | | Crystal structure of a beta-1,4-endoglucanase from Aspergillus niger | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, Y.J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-02-17 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Functional and structural analysis of Pichia pastoris-expressed Aspergillus niger 1,4-beta-endoglucanase
Biochem. Biophys. Res. Commun., 475, 2016
|
|
2P1W
 
 | |
3PYY
 
 | | Discovery and Characterization of a Cell-Permeable, Small-molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site | | Descriptor: | (5R)-5-[3-(4-fluorophenyl)-1-phenyl-1H-pyrazol-4-yl]imidazolidine-2,4-dione, 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, GLYCEROL, ... | | Authors: | Yang, J, Campobasso, N, Biju, M.P, Fisher, K, Pan, X.Q, Cottom, J, Galbraith, S, Ho, T, Zhang, H, Hong, X, Ward, P, Hofmann, G, Siegfried, B. | | Deposit date: | 2010-12-13 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and Characterization of a Cell-Permeable, Small-Molecule c-Abl Kinase Activator that Binds to the Myristoyl Binding Site.
Chem.Biol., 18, 2011
|
|
7FCA
 
 | | PfkB(Mycobacterium marinum) | | Descriptor: | Fructokinase, PfkB, GLYCEROL, ... | | Authors: | Li, J, Gao, B, Ji, R. | | Deposit date: | 2021-07-14 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural analysis and functional study of phosphofructokinase B (PfkB) from Mycobacterium marinum.
Biochem.Biophys.Res.Commun., 579, 2021
|
|
