2HDZ
 
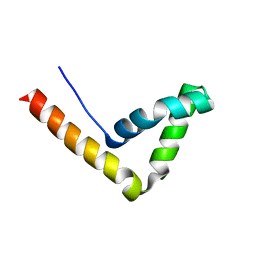 | | Crystal Structure Analysis of the UBF HMG box5 | | Descriptor: | Nucleolar transcription factor 1 | | Authors: | Rong, H, Teng, M.K, Niu, L.W. | | Deposit date: | 2006-06-21 | | Release date: | 2007-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human upstream binding factor HMG box 5 and site for binding of the cell-cycle regulatory factor TAF1
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5ZYA
 
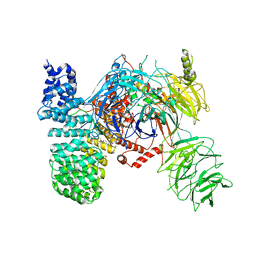 | | SF3b spliceosomal complex bound to E7107 | | Descriptor: | PHD finger-like domain-containing protein 5A, POTASSIUM ION, Splicing factor 3B subunit 1, ... | | Authors: | Finci, L.I, Larsen, N.A. | | Deposit date: | 2018-05-23 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action
Genes Dev., 32, 2018
|
|
6ACE
 
 | |
5X4R
 
 | | Structure of the N-terminal domain (NTD) of MERS-CoV spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S protein | | Authors: | Yuan, Y, Zhang, Y, Qi, J, Shi, Y, Gao, G.F. | | Deposit date: | 2017-02-14 | | Release date: | 2017-05-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains
Nat Commun, 8, 2017
|
|
2FKC
 
 | |
1LBA
 
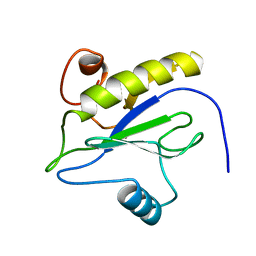 | |
2NOG
 
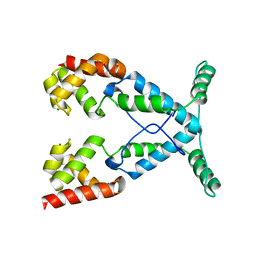 | |
4M5E
 
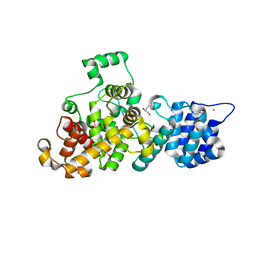 | | Tse3 structure | | Descriptor: | CADMIUM ION, CALCIUM ION, GLYCEROL, ... | | Authors: | Qian, Y. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
2FLC
 
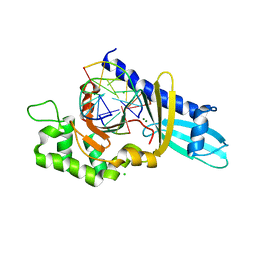 | |
5TJ8
 
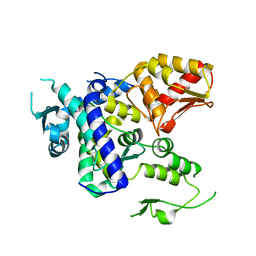 | |
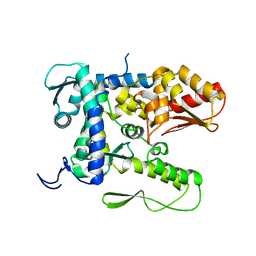
5TJ7
 
 | |
5TJQ
 
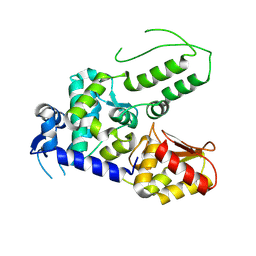 | | Structure of WWP2 2,3-linker-HECT | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2,NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Chen, Z, Gabelli, S.B. | | Deposit date: | 2016-10-04 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A Tunable Brake for HECT Ubiquitin Ligases.
Mol. Cell, 66, 2017
|
|
2JSO
 
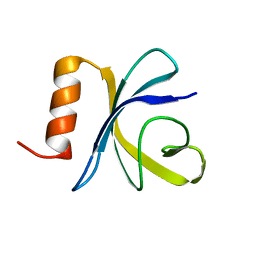 | | Antimicrobial resistance protein | | Descriptor: | Polymyxin resistance protein pmrD | | Authors: | Jin, C, Fu, W. | | Deposit date: | 2007-07-10 | | Release date: | 2007-09-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | First structure of the polymyxin resistance proteins.
Biochem.Biophys.Res.Commun., 361, 2007
|
|
4M5F
 
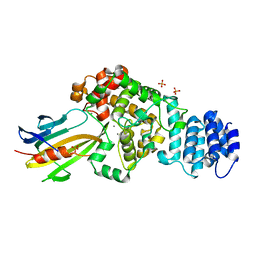 | | complex structure of Tse3-Tsi3 | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Uncharacterized protein | | Authors: | Gu, L.C. | | Deposit date: | 2013-08-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the T6SS effector protein Tse3 and the Tse3-Tsi3 complex from Pseudomonas aeruginosa reveal a calcium-dependent membrane-binding mechanism
Mol.Microbiol., 92, 2014
|
|
2FKH
 
 | |
6PIT
 
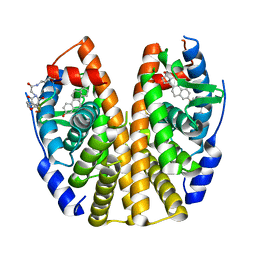 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with SRC2 Stapled Peptide 41A and Estradiol | | Descriptor: | ESTRADIOL, Estrogen receptor, Stapled Peptide 41A | | Authors: | Fanning, S.W, Montgomery, J.E, Greene, G.L, Moellering, R.E. | | Deposit date: | 2019-06-27 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Versatile Peptide Macrocyclization with Diels-Alder Cycloadditions.
J.Am.Chem.Soc., 141, 2019
|
|
8HPB
 
 | |
8HQB
 
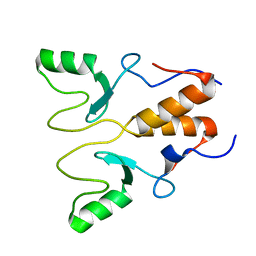 | | NMR Structure of OsCIE1-Ubox | | Descriptor: | U-box domain-containing protein 12 | | Authors: | Zhang, Y, Yu, C.Z, Lan, W.X. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Last modified: | 2024-06-12 | | Method: | SOLUTION NMR | | Cite: | Release of a ubiquitin brake activates OsCERK1-triggered immunity in rice.
Nature, 629, 2024
|
|
5GOU
 
 | | Structure of a 16-mer protein nanocage fabricated from its 24-mer analogue by subunit interface redesign | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, S, Zang, J, Wang, W, Wang, H, Zhao, G. | | Deposit date: | 2016-07-29 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Conversion of the Native 24-mer Ferritin Nanocage into Its Non-Native 16-mer Analogue by Insertion of Extra Amino Acid Residues.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
7CCF
 
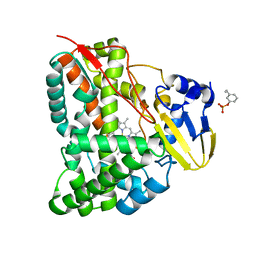 | | Mechanism insights on steroselective oxidation of phosphorylated ethylphenols with cytochrome P450 CreJ | | Descriptor: | (3-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S.Y, Feng, Y.G. | | Deposit date: | 2020-06-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Selective Oxidation of Phosphorylated Ethylphenols by Cytochrome P450 Monooxygenase CreJ.
Appl.Environ.Microbiol., 87, 2021
|
|
8JLP
 
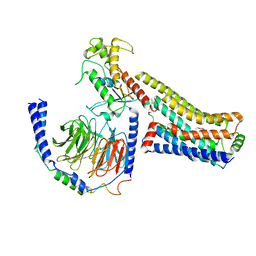 | | Ralmitaront(RO-6889450)-bound hTAAR1-Gs protein complex | | Descriptor: | 5-ethyl-4-methyl-~{N}-[4-[(2~{S})-morpholin-2-yl]phenyl]-1~{H}-pyrazole-3-carboxamide, Gs, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLK
 
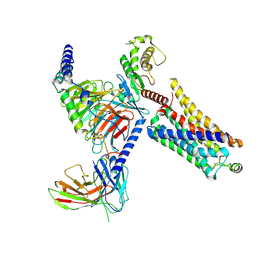 | | Ulotaront(SEP-363856)-bound mTAAR1-Gs protein complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLQ
 
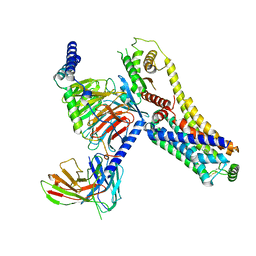 | | Fenoldopam-bound hTAAR1-Gs protein complex | | Descriptor: | (1R)-6-chloranyl-1-(4-hydroxyphenyl)-2,3,4,5-tetrahydro-1H-3-benzazepine-7,8-diol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8JLN
 
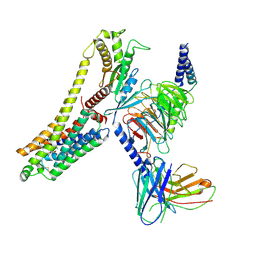 | | T1AM-bound hTAAR1-Gs protein complex | | Descriptor: | 4-[4-(2-azanylethyl)-2-iodanyl-phenoxy]phenol, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Xu, Z, Guo, L.L, Zhao, C, Shen, S.Y, Sun, J.P, Shao, Z.H. | | Deposit date: | 2023-06-02 | | Release date: | 2023-11-15 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Ligand recognition and G-protein coupling of trace amine receptor TAAR1.
Nature, 624, 2023
|
|
8FIV
 
 | |
