8ZLP
 
 | |
7BFZ
 
 | | X-ray structure of human prostate-specific membrane antigen(PSMA) in complex with a inhibitor Glu-490 | | Descriptor: | (((S)-1-carboxy-5-((E)-2-cyano-3-(5-(1-(3-methoxy-3-oxopropyl)-1,2,3,4-tetrahydroquinolin-6-yl)thiophen-2-yl)acrylamido)pentyl)carbamoyl)-L-glutamic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rakhimbekova, A, Motlova, L, Barinka, C. | | Deposit date: | 2021-01-05 | | Release date: | 2021-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A prostate-specific membrane antigen activated molecular rotor for real-time fluorescence imaging.
Nat Commun, 12, 2021
|
|
2FZP
 
 | | Crystal structure of the USP8 interaction domain of human NRDP1 | | Descriptor: | ring finger protein 41 isoform 1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Amino-terminal Dimerization, NRDP1-Rhodanese Interaction, and Inhibited Catalytic Domain Conformation of the Ubiquitin-specific Protease 8 (USP8).
J.Biol.Chem., 281, 2006
|
|
8ZDX
 
 | | Crystal structure of MjHKU4r-CoV-1 RBD bound to hDPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Yang, M, Li, Z, Xu, Y, Zhang, S. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for human DPP4 receptor recognition by a pangolin MERS-like coronavirus.
Plos Pathog., 20, 2024
|
|
8ZE6
 
 | | Crystal structure of MjHKU4r-CoV-1 RBD bound to MjDPP4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Yang, M, Li, Z, Xu, Y, Zhang, S. | | Deposit date: | 2024-05-04 | | Release date: | 2024-10-30 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for human DPP4 receptor recognition by a pangolin MERS-like coronavirus.
Plos Pathog., 20, 2024
|
|
5HC1
 
 | |
6O6L
 
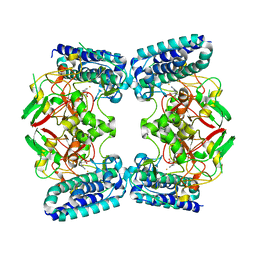 | | The Structure of EgtB(Cabther) in complex with Hercynine | | Descriptor: | EgtB (Cabther), FE (III) ION, N,N,N-trimethyl-histidine | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Ergothioneine Sulfoxide Synthase fromCandidatus Chloracidobacterium thermophilumand Structure-Guided Engineering To Modulate Its Substrate Selectivity.
Acs Catalysis, 9, 2019
|
|
8YQV
 
 | | African swine fever virus RNA Polymerase core | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Feng, X.Y. | | Deposit date: | 2024-03-20 | | Release date: | 2024-11-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Transcription regulation of African swine fever virus: dual role of M1249L.
Nat Commun, 15, 2024
|
|
8JFU
 
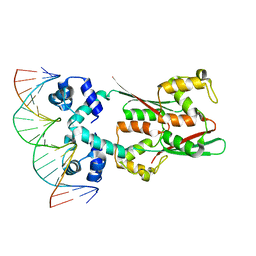 | | AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JFO
 
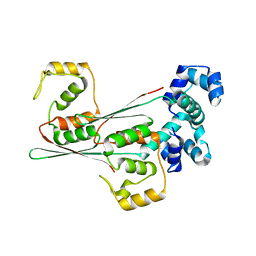 | |
8JFR
 
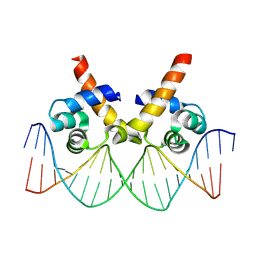 | | N-terminal domain of AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JG9
 
 | |
8JFT
 
 | | Cryo-EM structure of SaCas9-AcrIIA15 CTD-sgRNA complex | | Descriptor: | AcrIIA15, CRISPR-associated endonuclease Cas9, sgRNA of SaCas9 | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
6IRO
 
 | | the crosslinked complex of ISWI-nucleosome in the ADP-bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (167-MER), Histone H2A, ... | | Authors: | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | Deposit date: | 2018-11-13 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat. Struct. Mol. Biol., 26, 2019
|
|
5UN6
 
 | |
4HYP
 
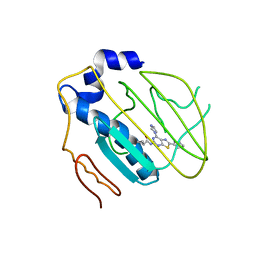 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | DNA gyrase subunit B, MAGNESIUM ION, N-[7-(1H-imidazol-1-yl)-2-(pyridin-3-yl)[1,3]thiazolo[5,4-d]pyrimidin-5-yl]cyclopropanecarboxamide | | Authors: | Bensen, D.C, Creighton, C.J, Tari, L.W. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HZ5
 
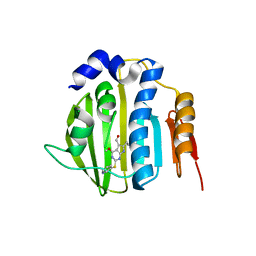 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity | | Descriptor: | 6-ethyl-4-methoxy-2-(pyridin-3-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbaldehyde, DNA topoisomerase IV, B subunit | | Authors: | Bensen, D.C, Creighton, C.J, Tari, L.W. | | Deposit date: | 2012-11-14 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HXZ
 
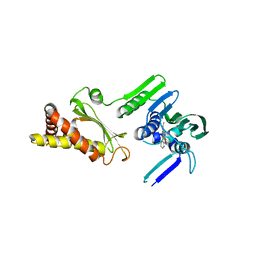 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 6-ethyl-4-methoxy-2-(pyridin-3-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbaldehyde, Topoisomerase IV, subunit B | | Authors: | Bensen, D.C, Creighton, C.J, Kwan, B, Tari, L.W. | | Deposit date: | 2012-11-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
5V7Q
 
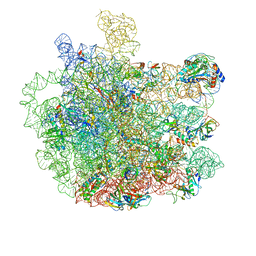 | | Cryo-EM structure of the large ribosomal subunit from Mycobacterium tuberculosis bound with a potent linezolid analog | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Yang, K, Chang, J.-Y, Cui, Z, Zhang, J. | | Deposit date: | 2017-03-20 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into species-specific features of the ribosome from the human pathogen Mycobacterium tuberculosis.
Nucleic Acids Res., 45, 2017
|
|
7VVE
 
 | | Complex structure of a leaf-branch compost cutinase variant in complex with mono(2-hydroxyethyl) terephthalic acid | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 4-(2-hydroxyethyloxycarbonyl)benzoic acid, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W44
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_RIP | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7VVC
 
 | | Crystal structure of inactive mutant of leaf-branch compost cutinase variant | | Descriptor: | ACETATE ION, ACETIC ACID, CALCIUM ION, ... | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-05 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W45
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KIP | | Descriptor: | CALCIUM ION, Leaf-branch compost cutinase, SODIUM ION | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-26 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
7W1N
 
 | | Complex structure of a leaf-branch compost cutinase variant LCC ICCG_KRP | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Leaf-branch compost cutinase | | Authors: | Niu, D, Zeng, W, Huang, J.W, Chen, C.C, Liu, W.D, Guo, R.T. | | Deposit date: | 2021-11-19 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substrate-Binding Mode of a Thermophilic PET Hydrolase and Engineering the Enzyme to Enhance the Hydrolytic Efficacy.
Acs Catalysis, 12, 2022
|
|
5V44
 
 | |
