8HAG
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (3.2 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAM
 
 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 2 | | Descriptor: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAH
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (3.9 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAI
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (4.7 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAN
 
 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 3 | | Descriptor: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAJ
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (4.8 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAK
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 4 (4.5 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
6L4T
 
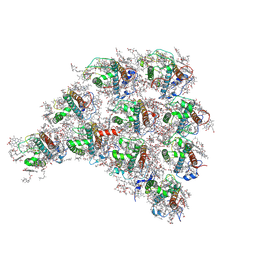 | | Structure of the peripheral FCPI from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Miyazaki, N, Akita, F, Shen, J.R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-20 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure of photosystem I-light-harvesting supercomplex from a red-lineage diatom
Nat Commun, 2020
|
|
6L4U
 
 | | Structure of the PSI-FCPI supercomplex from diatom | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Nagao, R, Kato, K, Miyazaki, N, Akita, F, Shen, J.R. | | Deposit date: | 2019-10-21 | | Release date: | 2020-05-20 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of photosystem I-light-harvesting supercomplex from a red-lineage diatom
Nat Commun, 2020
|
|
6LVL
 
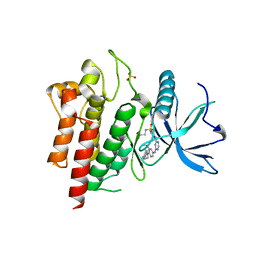 | | Crystal structure of FGFR2 in complex with 1,3,5-triazine derivative | | Descriptor: | Fibroblast growth factor receptor 2, N-ethyl-2-[[4-[[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]-1,3,5-triazin-2-yl]amino]benzenesulfonamide, SULFATE ION | | Authors: | Echizen, Y, Tateishi, Y, Amano, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2.
Bioorg.Med.Chem., 28, 2020
|
|
3ESW
 
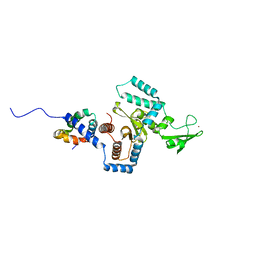 | | Complex of yeast PNGase with GlcNAc2-IAc. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase, UV excision repair protein RAD23, ... | | Authors: | Zhao, G, Zhou, X, Lennarz, W.J, Schindelin, H. | | Deposit date: | 2008-10-06 | | Release date: | 2008-11-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and mutational studies on the importance of oligosaccharide binding for the activity of yeast PNGase.
Glycobiology, 19, 2009
|
|
2RR6
 
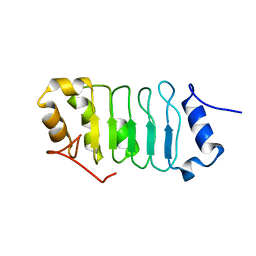 | | Solution structure of the leucine rich repeat of human acidic leucine-rich nuclear phosphoprotein 32 family member B | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B | | Authors: | Tochio, N, Umehara, T, Tsuda, K, Koshiba, S, Harada, T, Watanabe, S, Tanaka, A, Kigawa, T, Yokoyama, S. | | Deposit date: | 2010-05-25 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of histone chaperone ANP32B: interaction with core histones H3-H4 through its acidic concave domain.
J.Mol.Biol., 401, 2010
|
|
5FHI
 
 | | Crystallographic structure of PsoE without Co | | Descriptor: | GLUTATHIONE, Glutathione S-transferase, putative | | Authors: | Hara, K, Hashimoto, H, Yamamoto, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2015-12-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Oxidative trans to cis Isomerization of Olefins in Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5F8B
 
 | | Crystallographic Structure of PsoE with Co | | Descriptor: | (5~{S},8~{S},9~{R})-2-[(~{E})-hex-1-enyl]-8-methoxy-3-methyl-9-oxidanyl-8-(phenylcarbonyl)-1-oxa-7-azaspiro[4.4]non-2-ene-4,6-dione, COBALT (II) ION, GLUTATHIONE, ... | | Authors: | Hara, K, Hashimoto, H, Yamamoto, T, Tsunematsu, Y, Watanabe, K. | | Deposit date: | 2015-12-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Oxidative trans to cis Isomerization of Olefins in Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
5WXJ
 
 | | Apo EarP | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, GLYCEROL, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXI
 
 | | EarP bound with dTDP-rhamnose (soaked) | | Descriptor: | 2'-DEOXY-THYMIDINE-BETA-L-RHAMNOSE, BETA-MERCAPTOETHANOL, EarP, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
5WXK
 
 | | EarP bound with domain I of EF-P | | Descriptor: | BETA-MERCAPTOETHANOL, EarP, Elongation factor P, ... | | Authors: | Sengoku, T, Yokoyama, S, Yanagisawa, T. | | Deposit date: | 2017-01-07 | | Release date: | 2018-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural basis of protein arginine rhamnosylation by glycosyltransferase EarP
Nat. Chem. Biol., 14, 2018
|
|
1TLJ
 
 | | Crystal Structure of Conserved Protein of Unknown Function SSO0622 from Sulfolobus solfataricus | | Descriptor: | Hypothetical UPF0130 protein SSO0622, SULFATE ION | | Authors: | Jia, Z, Wong, A.H.Y, Kudrytska, M, Skarina, T, Walker, J, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-09 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and functional characterization of the TYW3/Taw3 class of SAM-dependent methyltransferases.
Rna, 23, 2017
|
|
6LVK
 
 | | Crystal structure of FGFR2 in complex with 1,3,5-triazine derivative | | Descriptor: | Fibroblast growth factor receptor 2, N-ethyl-2-[[4-[[4-(4-methylpiperazin-1-yl)-3-(2-morpholin-4-ylethoxy)phenyl]amino]-1,3,5-triazin-2-yl]amino]benzenesulfonamide, SULFATE ION | | Authors: | Echizen, Y, Amano, Y, Tateishi, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2.
Bioorg.Med.Chem., 28, 2020
|
|
6N30
 
 | | Bacillus PS3 ATP synthase class 3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6N2Y
 
 | | Bacillus PS3 ATP synthase class 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6N2D
 
 | | Bacillus PS3 ATP synthase membrane region | | Descriptor: | ATP synthase subunit a, ATP synthase subunit b, ATP synthase subunit c | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6N2Z
 
 | | Bacillus PS3 ATP synthase class 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Guo, H, Rubinstein, J.L. | | Deposit date: | 2018-11-14 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of a bacterial ATP synthase.
Elife, 8, 2019
|
|
6LVM
 
 | | Crystal structure of FGFR3 in complex with pyrimidine derivative | | Descriptor: | 2-[[5-[2-(3,5-dimethoxyphenyl)ethyl]-2-[[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]amino]pyrimidin-4-yl]amino]-N-ethyl-benzenesulfonamide, Fibroblast growth factor receptor 3 | | Authors: | Echizen, Y, Tateishi, Y, Amano, Y. | | Deposit date: | 2020-02-04 | | Release date: | 2020-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure-based drug design of 1,3,5-triazine and pyrimidine derivatives as novel FGFR3 inhibitors with high selectivity over VEGFR2.
Bioorg.Med.Chem., 28, 2020
|
|
8WZR
 
 | | SFX structure of an Mn-carbonyl complex immobilized in hen egg white lysozyme microcrystals, 100 ns after photoexcitation at RT. | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Maity, B, Shoji, M, Luo, F, Nakane, T, Abe, S, Owada, S, Kang, J, Tono, K, Tanaka, R, Thuc, T.T, Kojima, M, Hishikawa, Y, Tanaka, J, Tian, J, Noya, H, Nakasuji, Y, Asanuma, A, Yao, X, Iwata, S, Shigeta, Y, Nango, E, Ueno, T. | | Deposit date: | 2023-11-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Real-time observation of a metal complex-driven reaction intermediate using a porous protein crystal and serial femtosecond crystallography.
Nat Commun, 15, 2024
|
|
