8D6L
 
 | | Q108K:K40L:T51C:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V | | Descriptor: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, GLYCEROL, ... | | Authors: | Bingham, C.R, Geiger, J.H, Borhan, B. | | Deposit date: | 2022-06-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
4IMK
 
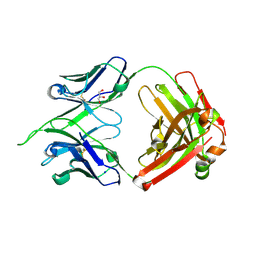 | | Uncrossed Fab binding to human Angiopoietin 2 | | Descriptor: | GLYCEROL, Heavy Chain, Light Chain, ... | | Authors: | Fenn, S, Schiller, C, Griese, J.J, Hopfner, K.-P, Kettenberger, H. | | Deposit date: | 2013-01-03 | | Release date: | 2013-04-17 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Crystal Structure of an Anti-Ang2 CrossFab Demonstrates Complete Structural and Functional Integrity of the Variable Domain.
Plos One, 8, 2013
|
|
4IML
 
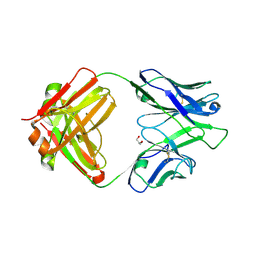 | | CrossFab binding to human Angiopoietin 2 | | Descriptor: | Crossed heavy chain (VH-Ckappa), Crossed light chain (VL-CH1), GLYCEROL | | Authors: | Fenn, S, Schiller, C, Griese, J, Hopfner, K.-P, Kettenberger, H. | | Deposit date: | 2013-01-03 | | Release date: | 2013-04-17 | | Last modified: | 2013-05-22 | | Method: | X-RAY DIFFRACTION (2.931 Å) | | Cite: | Crystal Structure of an Anti-Ang2 CrossFab Demonstrates Complete Structural and Functional Integrity of the Variable Domain.
Plos One, 8, 2013
|
|
8DN1
 
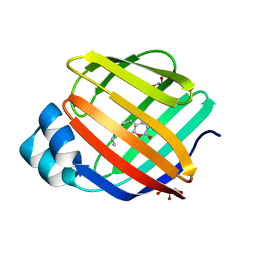 | | Q108K:K40L:T51C:T53A:R58L:Q38F:Q4F mutant of hCRBPII bound to synthetic fluorophore CM1V at pH 7.2 | | Descriptor: | (2E)-3-[7-(diethylamino)-2-oxo-2H-1-benzopyran-3-yl]prop-2-enal, bound form, GLYCEROL, ... | | Authors: | Bingham, C.R, Borhan, B, Geiger, J.H. | | Deposit date: | 2022-07-10 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Light controlled reversible Michael addition of cysteine: a new tool for dynamic site-specific labeling of proteins.
Analyst, 148, 2023
|
|
4M83
 
 | | Ensemble refinement of protein crystal structure (2IYF) of macrolide glycosyltransferases OleD complexed with UDP and Erythromycin A | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, Oleandomycin glycosyltransferase, ... | | Authors: | Wang, F, Helmich, K.E, Xu, W, Singh, S, Olmos Jr, J.L, Martinez iii, E, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-08-12 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Crystal structure of macrolide glycosyltransferases OleD
To be Published
|
|
6MCU
 
 | |
6MCV
 
 | |
6VIT
 
 | |
6MKV
 
 | |
5DPQ
 
 | | Crystal Structure of E72A mutant of domain swapped dimer Human Cellular Retinol Binding Protein | | Descriptor: | ACETATE ION, Retinol-binding protein 2 | | Authors: | Assar, Z, Nossoni, Z, Wang, W, Geiger, J.H, Borhan, B. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | Domain-Swapped Dimers of Intracellular Lipid-Binding Proteins: Evidence for Ordered Folding Intermediates.
Structure, 24, 2016
|
|
6VIS
 
 | |
6MLB
 
 | |
1RV1
 
 | | CRYSTAL STRUCTURE OF HUMAN MDM2 WITH AN IMIDAZOLINE INHIBITOR | | Descriptor: | CIS-[4,5-BIS-(4-BROMOPHENYL)-2-(2-ETHOXY-4-METHOXYPHENYL)-4,5-DIHYDROIMIDAZOL-1-YL]-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]METHANONE, Ubiquitin-protein ligase E3 Mdm2 | | Authors: | Lukacs, C, Kammlott, U, Graves, B. | | Deposit date: | 2003-12-12 | | Release date: | 2004-01-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In vivo activation of the p53 pathway by small-molecule antagonists of MDM2.
Science, 303, 2004
|
|
2IGK
 
 | | Crystal structure of recombinant pyranose 2-oxidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose oxidase | | Authors: | Divne, C. | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for substrate binding and regioselective oxidation of monosaccharides at c3 by pyranose 2-oxidase.
J.Biol.Chem., 281, 2006
|
|
2IGM
 
 | | Crystal structure of recombinant pyranose 2-oxidase H548N mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose oxidase | | Authors: | Divne, C. | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate binding and regioselective oxidation of monosaccharides at c3 by pyranose 2-oxidase.
J.Biol.Chem., 281, 2006
|
|
2IGN
 
 | | Crystal structure of recombinant pyranose 2-oxidase H167A mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose oxidase | | Authors: | Divne, C. | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for substrate binding and regioselective oxidation of monosaccharides at c3 by pyranose 2-oxidase.
J.Biol.Chem., 281, 2006
|
|
3TH9
 
 | | Crystal Structure of HIV-1 Protease Mutant Q7K V32I L63I with a cyclic sulfonamide inhibitor | | Descriptor: | Gag-Pol polyprotein, tert-butyl {(2S,3R)-4-[(4S)-7-fluoro-4-methyl-1,1-dioxido-4,5-dihydro-1,2-benzothiazepin-2(3H)-yl]-3-hydroxy-1-phenylbutan-2-yl}carbamate | | Authors: | Orth, P. | | Deposit date: | 2011-08-18 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Design, Synthesis, and X-ray Crystallographic Analysis of a Novel Class of HIV-1 Protease Inhibitors.
J.Med.Chem., 54, 2011
|
|
2IGO
 
 | |
6HRP
 
 | |
6HRT
 
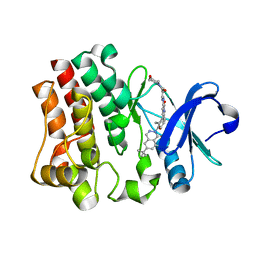 | | CRYSTAL STRUCTURE OF BTK KINASE DOMAIN COMPLEXED WITH 12-(6-tert-butyl-8-fluoro-1-oxo-phthalazin-2-yl)-9-hydroxy-6-methyl-4-[[5-(morpholine-4-carbonyl)-2-pyridyl]amino]-6-azatricyclo[9.4.0.02,7]pentadeca-1(15),2(7),3,11,13-pentaen-5-one | | Descriptor: | (9~{S})-12-(6-~{tert}-butyl-8-fluoranyl-1-oxidanylidene-phthalazin-2-yl)-6-methyl-4-[(5-morpholin-4-ylcarbonylpyridin-2-yl)amino]-9-oxidanyl-6-azatricyclo[9.4.0.0^{2,7}]pentadeca-1(15),2(7),3,11,13-pentaen-5-one, Tyrosine-protein kinase BTK | | Authors: | Kuglstatter, A, Janson, C. | | Deposit date: | 2018-09-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A potent seven-membered cyclic BTK (Bruton's tyrosine Kinase) chiral inhibitor conceived by structure-based drug design to lock its bioactive conformation.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
4DFF
 
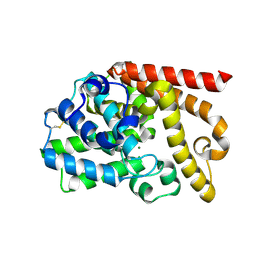 | | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia | | Descriptor: | 8,9-dimethoxy-1-(1,3-thiazol-5-yl)-5,6-dihydroimidazo[5,1-a]isoquinoline, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ho, G.D, Seganish, W.M, Bercovici, A, Tulshian, D, Greenlee, W.J, Van Rijn, R, Hruza, A, Xiao, L, Rindgen, D, Mullins, D, Guzzi, M, Zhang, X, Bleichardt, C, Hodgson, R. | | Deposit date: | 2012-01-23 | | Release date: | 2012-03-14 | | Last modified: | 2012-04-04 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The SAR development of dihydroimidazoisoquinoline derivatives as phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2FRS
 
 | |
2FDB
 
 | |
2FS6
 
 | |
6C7Z
 
 | | Crystal structure of the Q108K:K40L:T51V:R58F mutant of human Cellular Retinol Binding Protein II in complex with synthetic Ligand Julolidine | | Descriptor: | (2E,4E)-3-methyl-5-(2,3,6,7-tetrahydro-1H,5H-pyrido[3,2,1-ij]quinolin-9-yl)penta-2,4-dienal, ACETATE ION, Retinol-binding protein 2 | | Authors: | Nosrati, M, Geiger, J.H. | | Deposit date: | 2018-01-23 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | A Genetically Encoded Ratiometric pH Probe: Wavelength Regulation-Inspired Design of pH Indicators.
Chembiochem, 19, 2018
|
|
