7PS7
 
 | |
7Q0I
 
 | | Crystal structure of the N-terminal domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS5
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-47 Fab | | Descriptor: | Beta-47 Fab heavy chain, Beta-47 Fab light chain, Spike protein S1, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS1
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-27 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-27 Fab heavy chain, Beta-27 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7PS6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-44 and Beta-54 Fabs | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-44 Fab heavy chain, Beta-44 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0G
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-49 and FI-3A Fabs | | Descriptor: | Beta-49 Fab heavy chain, Beta-49 Fab light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q0H
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 beta variant spike glycoprotein in complex with Beta-50 and Beta-54 | | Descriptor: | Beta-50 Fab heavy chain, Beta-50 Fab light chain, Beta-54 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-10-14 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
8BH5
 
 | | SARS-CoV-2 BA.2.12.1 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | Beta-27 heavy chain, Beta-27 light chain, GLYCEROL, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Humoral responses against SARS-CoV-2 Omicron BA.2.11, BA.2.12.1 and BA.2.13 from vaccine and BA.1 serum.
Cell Discov, 8, 2022
|
|
8BBN
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-10 and EY6A Fabs | | Descriptor: | BA.2-10 heavy chain, BA.2-10 light chain, EY6A Heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8BBO
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-36 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IGH@ protein, Immunoglobulin kappa light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
8BCZ
 
 | | SARS-CoV-2 Delta-RBD complexed with Fabs BA.2-36, BA.2-23, EY6A and COVOX-45 | | Descriptor: | BA.2-23 heavy chain, BA.2-23 light chain, BA.2-36 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
7U0K
 
 | | IOMA class antibody Fab ACS124 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, IOMA Class antibody ACS124 Heavy chain, IOMA Class antibody ACS124 Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U04
 
 | | IOMA class antibody ACS101 | | Descriptor: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U5G
 
 | | ACS122 Fab | | Descriptor: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
6B70
 
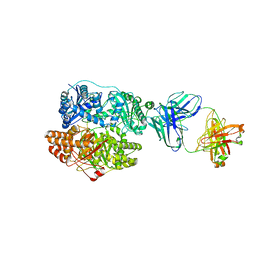 | | Cryo-EM structure of human insulin degrading enzyme in complex with FAB H11-E heavy chain, FAB H11-E light chain and insulin | | Descriptor: | FAB H11-E heavy chain, FAB H11-E light chain, Insulin, ... | | Authors: | Liang, W.G, Zhang, Z, Bailey, L.J, Kossiakoff, A.A, Tan, Y.Z, Wei, H, Carragher, B, Potter, S.C, Tang, W.J. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
7Z3A
 
 | | AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACS101 heavy chain, ... | | Authors: | van Schooten, J, Ward, A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7Q9P
 
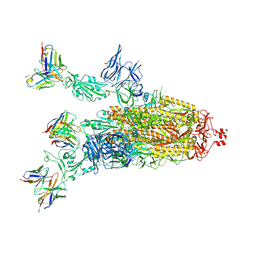 | | Beta-06 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-06 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9F
 
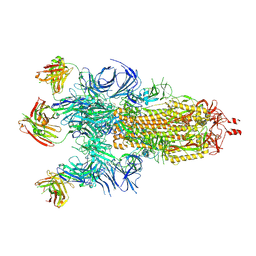 | | Beta-50 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9J
 
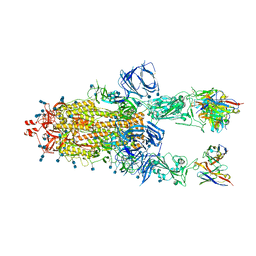 | | Beta-26 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-26 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q6E
 
 | | Beta049 fab in complex with SARS-CoV2 beta-Spike glycoprotein, The Beta mAb response underscores the antigenic distance to other SARS-CoV-2 variants | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-49 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-07 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9G
 
 | | COVOX-222 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9I
 
 | | Beta-43 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-43 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9K
 
 | | Beta-32 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-32 heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7Q9M
 
 | | Beta-53 fab in complex with SARS-CoV-2 beta-Spike glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-53 fab heavy chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2021-11-12 | | Release date: | 2021-12-15 | | Last modified: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The antibody response to SARS-CoV-2 Beta underscores the antigenic distance to other variants.
Cell Host Microbe, 30, 2022
|
|
7ZLK
 
 | | AMC009 SOSIPv5.2 in complex with Fabs ACS114 and ACS122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACS114 heavy chain, ... | | Authors: | van Schooten, J, Ozorowski, G, Ward, A. | | Deposit date: | 2022-04-15 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
