7WTV
 
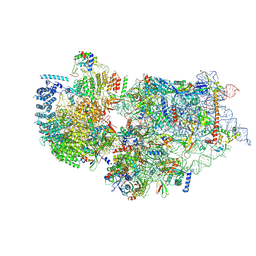 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A2 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7WTW
 
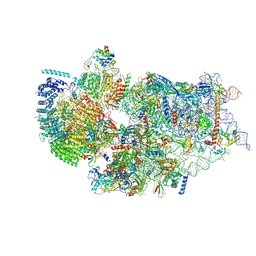 | | Cryo-EM structure of a human pre-40S ribosomal subunit - State RRP12-A3 | | Descriptor: | 18S rRNA, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
7CTG
 
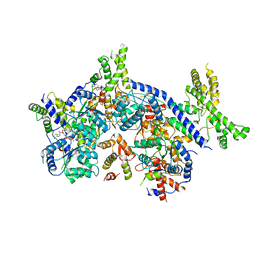 | | Human Origin Recognition Complex, ORC1-5 State I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
7CTE
 
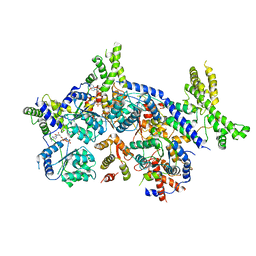 | | Human Origin Recognition Complex, ORC2-5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 2, Origin recognition complex subunit 3, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
7CTF
 
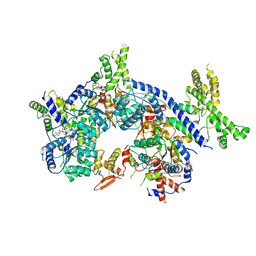 | | Human origin recognition complex 1-5 State II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
7Y54
 
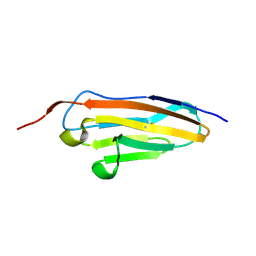 | |
7Y6O
 
 | |
7Y9A
 
 | | Crystal structure of sDscam Ig1-2 domains, isoform beta2v6 | | Descriptor: | Down Syndrome Cell Adhesion Molecules, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-3)][beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y5J
 
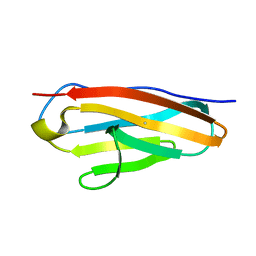 | |
7Y5R
 
 | |
7Y73
 
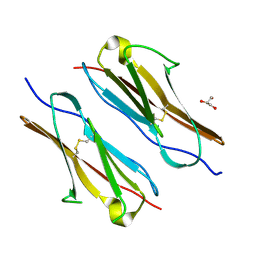 | |
7Y6E
 
 | |
7Y8S
 
 | |
7Y8I
 
 | | Crystal structure of sDscam FNIII3 domain, isoform alpha7 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Dscam, ... | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y8H
 
 | |
7Y95
 
 | | Crystal structure of sDscam Ig1 domain, isoform beta6v2 | | Descriptor: | Dscam, GLYCEROL, SODIUM ION | | Authors: | Chen, Q, Yu, Y, Cheng, J. | | Deposit date: | 2022-06-24 | | Release date: | 2023-05-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the self-recognition of sDSCAM in Chelicerata.
Nat Commun, 14, 2023
|
|
7Y4X
 
 | |
4XAX
 
 | | Crystal structure of Thermus thermophilus CarD in complex with the Thermus aquaticus RNA polymerase beta1 domain | | Descriptor: | 1,2-ETHANEDIOL, CarD, DNA-directed RNA polymerase subunit beta domain 1 | | Authors: | Chen, J, Bae, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | CarD uses a minor groove wedge mechanism to stabilize the RNA polymerase open promoter complex.
Elife, 4, 2015
|
|
3LDA
 
 | | Yeast Rad51 H352Y Filament Interface Mutant | | Descriptor: | CHLORIDE ION, DNA repair protein RAD51 | | Authors: | Villanueva, N.L, Chen, J, Morrical, S.W, Rould, M.A. | | Deposit date: | 2010-01-12 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the mechanism of Rad51 recombinase from the structure and properties of a filament interface mutant.
Nucleic Acids Res., 38, 2010
|
|
8KAG
 
 | | Crystal structure of SpyCas9 in complex with sgRNA and target RNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, RNA (5'-R(*CP*CP*AP*CP*UP*UP*CP*AP*AP*UP*UP*AP*GP*AP*AP*CP*AP*CP*GP*GP*AP*CP*C)-3'), RNA (98-MER) | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.88 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 2024
|
|
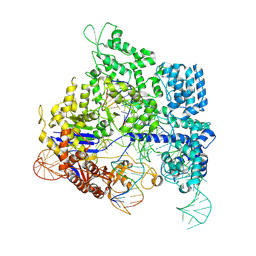
8KAK
 
 | |
8KAJ
 
 | | Crystal structure of SpyCas9-crRNA-tracrRNA complex bound to 16nt target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*CP*AP*AP*TP*AP*CP*CP*TP*TP*TP*TP*AP*TP*CP*CP*AP*TP*AP*AP*AP*TP*TP*CP*G)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*GP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 2024
|
|
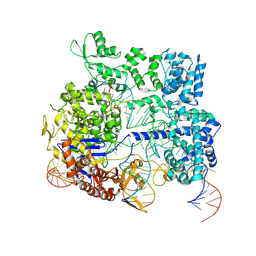
8KAI
 
 | |
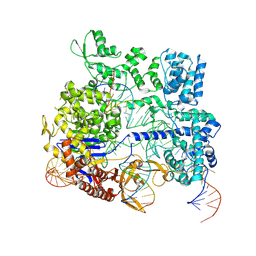
8KAH
 
 | |
8KAM
 
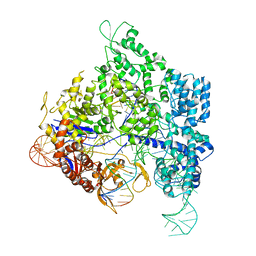 | | Crystal structure of SpyCas9 in complex with sgRNA and 16nt target DNA | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*CP*AP*AP*TP*AP*CP*CP*TP*CP*TP*TP*CP*AP*AP*TP*TP*AP*GP*AP*AP*CP*AP*CP*G)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*GP*TP*AP*TP*TP*G)-3'), ... | | Authors: | Chen, Y, Chen, J, Liu, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | Trans-nuclease activity of Cas9 activated by DNA or RNA target binding.
Nat.Biotechnol., 2024
|
|
