2RFQ
 
 | | Crystal structure of 3-HSA hydroxylase from Rhodococcus sp. RHA1 | | Descriptor: | 3-HSA hydroxylase, oxygenase, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE | | Authors: | Chang, C, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-01 | | Release date: | 2007-10-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of 3-HSA hydroxylase, oxygenase from Rhodococcus sp. RHA1.
To be Published
|
|
2RA5
 
 | | Crystal structure of the putative transcriptional regulator from Streptomyces coelicolor | | Descriptor: | ISOPROPYL ALCOHOL, Putative transcriptional regulator, S,R MESO-TARTARIC ACID | | Authors: | Kim, Y, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the putative transcriptional regulator from Streptomyces coelicolor.
To be Published
|
|
2RAE
 
 | | Crystal structure of a TetR/AcrR family transcriptional regulator from Rhodococcus sp. RHA1 | | Descriptor: | Transcriptional regulator, AcrR family protein | | Authors: | Zhang, R, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of a TetR/AcrR family transcriptional regulator from Rhodococcus sp. RHA1
To be Published
|
|
2RBC
 
 | | Crystal structure of a putative ribokinase from Agrobacterium tumefaciens | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cuff, M.E, Xu, X, Zheng, H, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-18 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a putative ribokinase from Agrobacterium tumefaciens.
TO BE PUBLISHED
|
|
1JQ3
 
 | | Crystal Structure of Spermidine Synthase in Complex with Transition State Analogue AdoDATO | | Descriptor: | S-ADENOSYL-1,8-DIAMINO-3-THIOOCTANE, Spermidine synthase | | Authors: | Korolev, S, Ikeguchi, Y, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Pegg, A.E, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-08-03 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of spermidine synthase with a multisubstrate adduct inhibitor.
Nat.Struct.Biol., 9, 2002
|
|
2R6I
 
 | | Crystal structure of Atu1473 protein, a putative chaperone from Agrobacterium tumefaciens | | Descriptor: | Uncharacterized protein Atu1473 | | Authors: | Osipiuk, J, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-05 | | Release date: | 2007-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of Atu1473 protein, a putative chaperone from Agrobacterium tumefaciens.
To be Published
|
|
2R6U
 
 | | Crystal structure of gene product RHA04853 from Rhodococcus sp. RHA1 | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, R, Xu, X, Gu, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-06 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of gene product RHA04853 from Rhodococcus sp. RHA1.
To be Published
|
|
1TU9
 
 | | Crystal Structure of a Protein PA3967, a Structurally Highly Homologous to a Human Hemoglobin, from Pseudomonas aeruginosa PAO1 | | Descriptor: | 1,2-ETHANEDIOL, PROPANOIC ACID, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Kim, Y, Joachimiak, A, Skarina, T, Egorova, O, Bochkarev, A, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-06-24 | | Release date: | 2004-08-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of PA3967 from Pseudomonas aeruginosa PAO1, a Hypothetical Protein which is highly homologous to human Hemoglobin in structure.
To be Published
|
|
2R8B
 
 | | The crystal structure of the protein Atu2452 of unknown function from Agrobacterium tumefaciens str. C58 | | Descriptor: | SULFATE ION, Uncharacterized protein Atu2452 | | Authors: | Tan, K, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-10 | | Release date: | 2007-09-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The crystal structure of the protein Atu2452 of unknown function from Agrobacterium tumefaciens str. C58.
TO BE PUBLISHED
|
|
1K4N
 
 | | Structural Genomics, Protein EC4020 | | Descriptor: | Protein EC4020 | | Authors: | Zhang, R.G, Joachimiak, A, Edwards, A, Savchenko, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-08 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved protein YecM from Escherichia coli shows structural homology to metal-binding isomerases and oxygenases.
Proteins, 51, 2003
|
|
8DVC
 
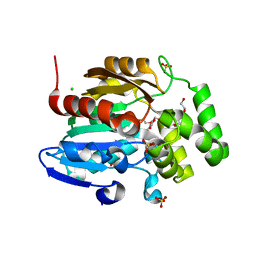 | | Receptor ShHTL5 from Striga hermonthica in complex with strigolactone agonist GR24 | | Descriptor: | (3R,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methyl)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Arellano-Saab, A, Skarina, T, Yim, V, Savchenko, A, Stogios, P.J, McCourt, P. | | Deposit date: | 2022-07-28 | | Release date: | 2023-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.638 Å) | | Cite: | Structural analysis of a hormone-bound Striga strigolactone receptor.
Nat.Plants, 9, 2023
|
|
5F13
 
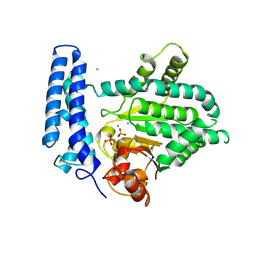 | | Structure of Mn bound DUF89 from Saccharomyces cerevisiae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Nocek, B, Skarina, T, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | A family of metal-dependent phosphatases implicated in metabolite damage-control.
Nat.Chem.Biol., 12, 2016
|
|
2RFL
 
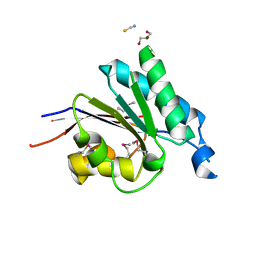 | | Crystal structure of the putative phosphohistidine phosphatase SixA from Agrobacterium tumefaciens | | Descriptor: | ACETIC ACID, GLYCEROL, Putative phosphohistidine phosphatase SixA, ... | | Authors: | Kim, Y, Binkowski, T, Xu, X, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-10-01 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Putative Phosphohistidine Phosphatase SixA from
Agrobacterium tumefaciens.
To be Published
|
|
6YNU
 
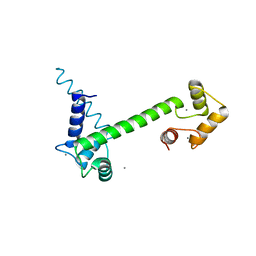 | | CaM-P458 complex (crystal form 1) | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Voegele, A, Haouz, A, Chenal, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells.
Adv Sci, 8, 2021
|
|
6YNS
 
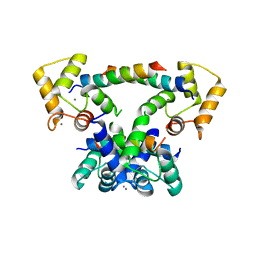 | | CaM-P458 complex (crystal form 2) | | Descriptor: | Bifunctional adenylate cyclase toxin/hemolysin CyaA, CALCIUM ION, Calmodulin-1 | | Authors: | Mechaly, A.E, Voegele, A, Haouz, A, Chenal, A. | | Deposit date: | 2020-04-14 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | A High-Affinity Calmodulin-Binding Site in the CyaA Toxin Translocation Domain is Essential for Invasion of Eukaryotic Cells.
Adv Sci, 8, 2021
|
|
5IBZ
 
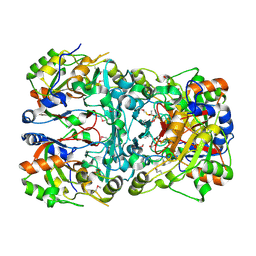 | | Crystal structure of a novel cyclase (pfam04199). | | Descriptor: | ACETYLPHOSPHATE, TRIETHYLENE GLYCOL, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Brown, G, Joachimiak, A, Savchenko, A, Yakunin, A. | | Deposit date: | 2016-02-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | Crystal structure of a novel cyclase (pfam04199).
To Be Published
|
|
3LG2
 
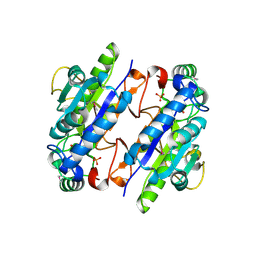 | | A Ykr043C/ fructose-1,6-bisphosphate product complex following ligand soaking | | Descriptor: | PHOSPHATE ION, Uncharacterized protein YKR043C | | Authors: | Singer, A, Xu, X, Cui, H, Dong, A, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-19 | | Release date: | 2010-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and activity of the metal-independent fructose-1,6-bisphosphatase YK23 from Saccharomyces cerevisiae.
J.Biol.Chem., 285, 2010
|
|
1H2H
 
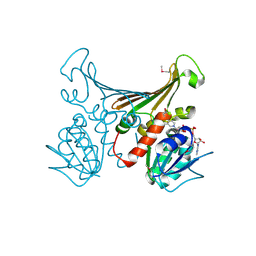 | | Crystal structure of TM1643 | | Descriptor: | HYPOTHETICAL PROTEIN TM1643, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yang, Z, Savchenko, A, Edwards, A, Arrowsmith, C, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-08 | | Release date: | 2002-08-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aspartate dehydrogenase, a novel enzyme identified from structural and functional studies of TM1643.
J. Biol. Chem., 278, 2003
|
|
7SNU
 
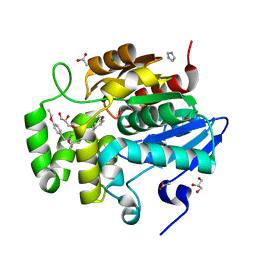 | | Crystal structure of ShHTL7 from Striga hermonthica in complex with strigolactone antagonist RG6 | | Descriptor: | 2-{(2S)-1-[(4-ethoxyphenyl)methyl]-4-[(2E)-3-(4-methoxyphenyl)prop-2-en-1-yl]piperazin-2-yl}ethan-1-ol, ACETATE ION, GLYCEROL, ... | | Authors: | Arellano-Saab, A, Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, McCourt, P. | | Deposit date: | 2021-10-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A novel strigolactone receptor antagonist provides insights into the structural inhibition, conditioning, and germination of the crop parasite Striga.
J.Biol.Chem., 298, 2022
|
|
5IJI
 
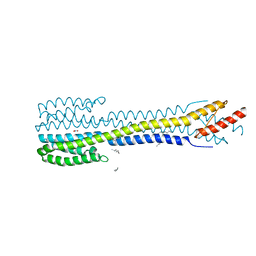 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (WT) in symmetric holo state | | Descriptor: | EICOSANE, NITRATE ION, Nitrate/nitrite sensor histidine kinase NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-03-02 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
5JEF
 
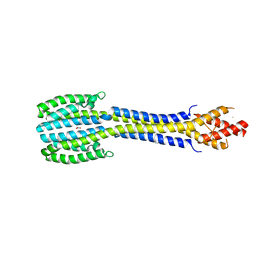 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (WT) in asymmetric holo state | | Descriptor: | EICOSANE, NITRATE ION, Nitrate/nitrite sensor protein NarQ | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5WP0
 
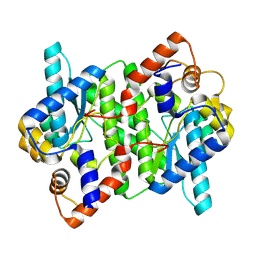 | | Crystal structure of NAD synthetase NadE from Vibrio fischeri | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Stogios, P.J, Evdokimova, E, Grimshaw, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-08-03 | | Release date: | 2017-08-16 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics of bacterial drug targets: Application of a high-throughput pipeline to solve 58 protein structures from pathogenic and related bacteria.
Microbiol Resour Announc, 2025
|
|
5JEQ
 
 | | Fragment of nitrate/nitrite sensor histidine kinase NarQ (R50K) in symmetric apo state | | Descriptor: | Nitrate/nitrite sensor protein NarQ, PHOSPHATE ION | | Authors: | Gushchin, I, Melnikov, I, Polovinkin, V, Ishchenko, A, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of transmembrane signaling by sensor histidine kinases.
Science, 356, 2017
|
|
1KXJ
 
 | | The Crystal Structure of Glutamine Amidotransferase from Thermotoga maritima | | Descriptor: | Amidotransferase hisH, PHOSPHATE ION | | Authors: | Korolev, S, Skarina, T, Evdokimova, E, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-31 | | Release date: | 2002-03-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of glutamine amidotransferase from Thermotoga maritima
Proteins, 49, 2002
|
|
