3FAQ
 
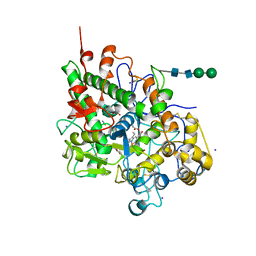 | | Crystal structure of lactoperoxidase complex with cyanide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CYANIDE ION, ... | | Authors: | Sheikh, I.A, Singh, N, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2008-11-18 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Evidence of Substrate Specificity in Mammalian Peroxidases: STRUCTURE OF THE THIOCYANATE COMPLEX WITH LACTOPEROXIDASE AND ITS INTERACTIONS AT 2.4 A RESOLUTION
J.Biol.Chem., 284, 2009
|
|
3MME
 
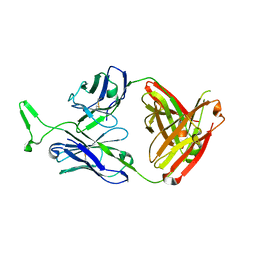 | | Structure and functional dissection of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PG16 HEAVY CHAIN FAB, ... | | Authors: | Pancera, M, McLellan, J, Zhou, T, Zhu, J, Kwong, P. | | Deposit date: | 2010-04-19 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.97 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
4GRH
 
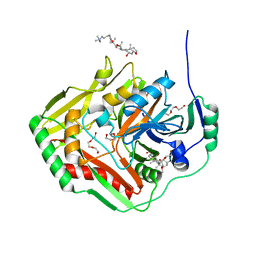 | | Crystal structure of pabB of Stenotrophomonas maltophilia | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, MAGNESIUM ION, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Bera, A, Atanasova, V, Ladner, J.E, Parsons, J.F. | | Deposit date: | 2012-08-24 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of Aminodeoxychorismate Synthase from Stenotrophomonas maltophilia.
Biochemistry, 51, 2012
|
|
5GH0
 
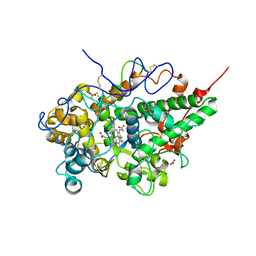 | | Crystal structure of the complex of bovine lactoperoxidase with mercaptoimidazole at 2.3 A resolution | | Descriptor: | 1,3-dihydroimidazole-2-thione, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Singh, P.K, Sirohi, H.V, Singh, A.K, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2016-06-17 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design of anti-thyroid drugs: Binding studies and structure determination of the complex of lactoperoxidase with 2-mercaptoimidazole at 2.30 angstrom resolution
Proteins, 85, 2017
|
|
7WY5
 
 | | ADGRL3/Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
7WYB
 
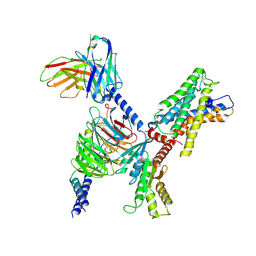 | | ADGRL3/Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and G q , G s , G i , and G 12 coupling.
Mol.Cell, 82, 2022
|
|
7WY8
 
 | | ADGRL3/Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-15 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling
Mol.Cell, 82, 2022
|
|
7X10
 
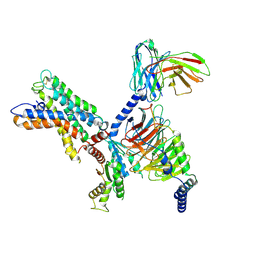 | | ADGRL3/miniG12 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Isoform 3 of Adhesion G protein-coupled receptor L3, ... | | Authors: | He, Y, Qian, Y. | | Deposit date: | 2022-02-22 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into adhesion GPCR ADGRL3 activation and Gq, Gs, Gi, and G12 coupling.
Mol.Cell, 82, 2022
|
|
9EQH
 
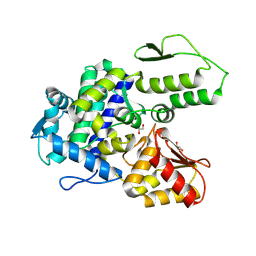 | | WWP2 WW2-2,3-linker-HECT (WWP2-LH) | | Descriptor: | GLYCEROL, Isoform 2 of NEDD4-like E3 ubiquitin-protein ligase WWP2, SODIUM ION | | Authors: | Dudey, A.P, Hemmings, A.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expanding the Inhibitor Space of the WWP1 and WWP2 HECT E3 Ligases
To Be Published
|
|
1V1P
 
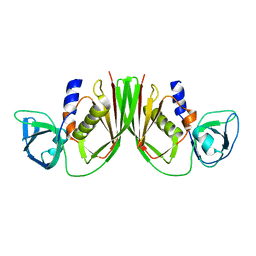 | |
4ALN
 
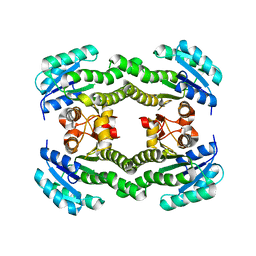 | | Crystal structure of S. aureus FabI (P32) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH] | | Authors: | Schiebel, J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
4ALM
 
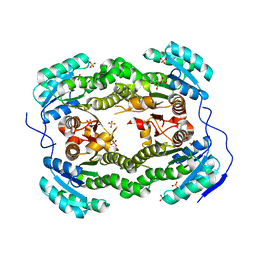 | | Crystal structure of S. aureus FabI (P43212) | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADPH], SULFATE ION | | Authors: | Schiebel, J, Kisker, C. | | Deposit date: | 2012-03-04 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Staphylococcus Aureus Fabi: Inhibition, Substrate Recognition and Potential Implications for in Vivo Essentiality
Structure, 20, 2012
|
|
3LRS
 
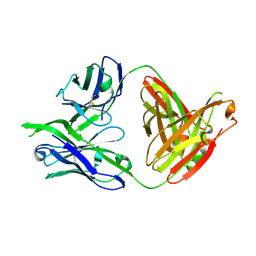 | | Structure of PG16, an antibody with broad and potent neutralization of HIV-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PG-16 Heavy Chain Fab, PG-16 Light Chain Fab | | Authors: | Pancera, M, Kwong, P.D. | | Deposit date: | 2010-02-11 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of PG16 and chimeric dissection with somatically related PG9: structure-function analysis of two quaternary-specific antibodies that effectively neutralize HIV-1.
J.Virol., 84, 2010
|
|
2HPG
 
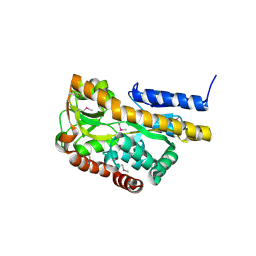 | |
2L9V
 
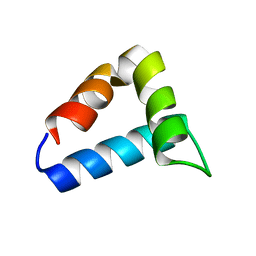 | | NMR structure of the FF domain L24A mutant's folding transition state | | Descriptor: | Pre-mRNA-processing factor 40 homolog A | | Authors: | Korzhnev, D.M, Vernon, R.M, Religa, T.L, Hansen, A, Baker, D, Fersht, A.R, Kay, L.E. | | Deposit date: | 2011-02-24 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nonnative interactions in the FF domain folding pathway from an atomic resolution structure of a sparsely populated intermediate: an NMR relaxation dispersion study.
J.Am.Chem.Soc., 133, 2011
|
|
4G9O
 
 | |
2DUJ
 
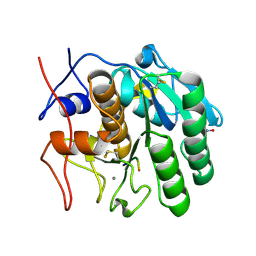 | | Crystal structure of the complex formed between proteinase K and a synthetic peptide Leu-Leu-Phe-Asn-Asp at 1.67 A resolution | | Descriptor: | CALCIUM ION, LLFND, NITRATE ION, ... | | Authors: | Singh, A.K, Singh, N, Somvanshi, R.K, Gupta, D, Sharma, S, Singh, T.P. | | Deposit date: | 2006-07-23 | | Release date: | 2006-08-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the complex of proteinase K with a specific lactoferrin peptide Val-Leu-Leu-His at 1.93 A resolution
To be Published
|
|
4M3L
 
 | | Crystal Structure of the coiled coil domain of MuRF1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, E3 ubiquitin-protein ligase TRIM63, ... | | Authors: | Mayans, O, Franke, B. | | Deposit date: | 2013-08-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the fold organization and sarcomeric targeting of the muscle atrogin MuRF1.
Open Biol, 4, 2014
|
|
4BHT
 
 | | Structural Determinants of Cofactor Specificity and Domain Flexibility in Bacterial Glutamate Dehydrogenases | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, NADP-SPECIFIC GLUTAMATE DEHYDROGENASE, ... | | Authors: | Oliveira, T, Sharkey, M, Engel, P, Khan, A. | | Deposit date: | 2013-04-06 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Nadp(+) -Dependent Glutamate Dehydrogenase from Escherichia Coli: Reflections on the Basis of Coenzyme Specificity in the Family of Glutamate Dehydrogenases.
FEBS J., 280, 2013
|
|
5O7T
 
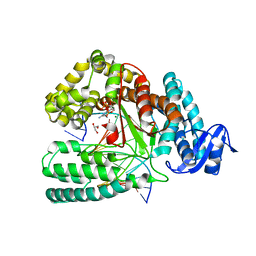 | | Crystal structure of KlenTaq mutant M747K in a closed ternary complex with a dG:dCTP base pair | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA polymerase I, thermostable, ... | | Authors: | Betz, K, Diederichs, K, Marx, A. | | Deposit date: | 2017-06-09 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the selective incorporation of an artificial nucleotide opposite a DNA adduct by a DNA polymerase.
Chem. Commun. (Camb.), 53, 2017
|
|
2C0R
 
 | |
3SEZ
 
 | |
5MF2
 
 | | Bacteriophage T5 distal tail protein pb9 co-crystallized with Tb-Xo4 | | Descriptor: | CHLORIDE ION, Distal tail protein, TERBIUM(III) ION, ... | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Lassalle, L, Arnaud, C.-A, Breyton, C, Madern, D, Coquelle, N, Maury, O, Girard, E. | | Deposit date: | 2016-11-17 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallophore: a versatile lanthanide complex for protein crystallography combining nucleating effects, phasing properties, and luminescence.
Chem Sci, 8, 2017
|
|
2EHA
 
 | | Crystal structure of goat lactoperoxidase complexed with formate anion at 3.3 A resolution | | Descriptor: | 1-(OXIDOSULFANYL)METHANAMINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Singh, A.K, Ethayathulla, A.S, Singh, N, Sharma, S, Kaur, P, Singh, T.P. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of goat lactoperoxidase complexed with formate anion at 3.3 A resolution
to be published
|
|
2BCJ
 
 | |
