4JX1
 
 | | Crystal structure of reduced Cytochrome P450cam-putidaredoxin complex bound to camphor and 5-exo-hydroxycamphor | | Descriptor: | 1,1'-hexane-1,6-diyldipyrrolidine-2,5-dione, 5-EXO-HYDROXYCAMPHOR, CALCIUM ION, ... | | Authors: | Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Structural basis for effector control and redox partner recognition in cytochrome P450.
Science, 340, 2013
|
|
3QOO
 
 | | Crystal structure of hot-dog-like Taci_0573 protein from Thermanaerovibrio acidaminovorans | | Descriptor: | CHLORIDE ION, SODIUM ION, Uncharacterized protein | | Authors: | Michalska, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-10 | | Release date: | 2011-02-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of hot-dog-like Taci_0573 protein from Thermanaerovibrio acidaminovorans
TO BE PUBLISHED
|
|
4NTP
 
 | |
4H4K
 
 | | Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA-silencing complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cmr subunit Cmr2, CRISPR system Cmr subunit Cmr3, ... | | Authors: | Shao, Y, Cocozaki, A.I, Ramia, N.F, Terns, R.M, Terns, M.P, Li, H. | | Deposit date: | 2012-09-17 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.804 Å) | | Cite: | Structure of the cmr2-cmr3 subcomplex of the cmr RNA silencing complex.
Structure, 21, 2013
|
|
4HD1
 
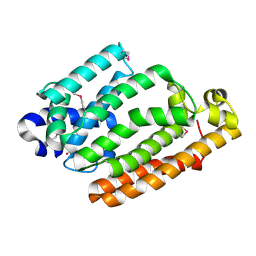 | |
5EGM
 
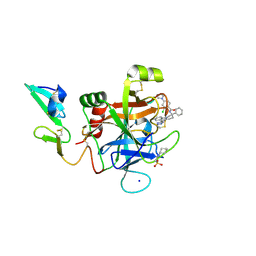 | | Development of a novel tricyclic class of potent and selective FIXa inhibitors | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, 2-chloranyl-~{N}-[(7~{S})-2-methyl-7-phenyl-10-(1~{H}-1,2,3,4-tetrazol-5-yl)-8,9-dihydro-6~{H}-pyrido[1,2-a]indol-7-yl]-4-(1,2,4-triazol-4-yl)benzamide, Coagulation factor IX, ... | | Authors: | Meng, D, Andre, P, Bateman, T.J, Berger, R, Chen, Y, Desai, K, Dewnani, S, Ellsworth, K, Feng, D, Geissler, W.M, Guo, L, Hruza, A, Jian, T, Li, H, Parker, D.L, Reichert, P, Sherer, E.C, Smith, C.J, Sonatore, L.M, Tschirret-Guth, R, Wu, J, Xu, J, Zhang, T, Campeau, L, Orr, R, Poirier, M, McCabe-Dunn, j, Araki, K, Nishimura, T, Sakurada, I, Hirabayashi, T, Wood, H.B. | | Deposit date: | 2015-10-27 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Development of a novel tricyclic class of potent and selective FIXa inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2BID
 
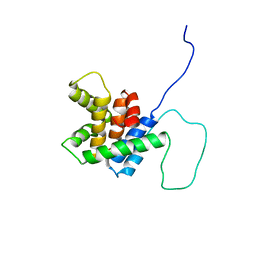 | | HUMAN PRO-APOPTOTIC PROTEIN BID | | Descriptor: | PROTEIN (BID) | | Authors: | Chou, J.J, Li, H, Salvesen, G.S, Yuan, J, Wagner, G. | | Deposit date: | 1999-01-27 | | Release date: | 2000-02-02 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BID, an intracellular amplifier of apoptotic signaling.
Cell(Cambridge,Mass.), 96, 1999
|
|
5EGG
 
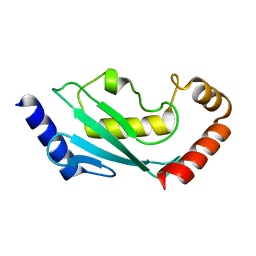 | |
4JTS
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 072 | | Descriptor: | 4-methyl-N-(naphthalen-2-yl)-1,3-thiazole-5-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Zhu, L, Zhu, J, Ren, X, Li, H. | | Deposit date: | 2013-03-24 | | Release date: | 2014-03-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with 057
To be Published
|
|
4MVE
 
 | |
4MZG
 
 | | Crystal structure of human Spindlin1 bound to histone H3K4me3 peptide | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Su, X, Ding, X, Li, H. | | Deposit date: | 2013-09-30 | | Release date: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1
Genes Dev., 28, 2014
|
|
4NAS
 
 | | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | CALCIUM ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The crystal structure of a rubisco-like protein (MtnW) from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446.
To be Published
|
|
4MZF
 
 | | Crystal structure of human Spindlin1 bound to histone H3(K4me3-R8me2a) peptide | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Peptide from Histone H3.2, ... | | Authors: | Su, X, Ding, X, Li, H. | | Deposit date: | 2013-09-30 | | Release date: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1
Genes Dev., 28, 2014
|
|
4N03
 
 | | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata | | Descriptor: | 1,2-ETHANEDIOL, ABC-type branched-chain amino acid transport systems periplasmic component-like protein, PALMITIC ACID | | Authors: | Osipiuk, J, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Fatty acid ABC transporter substrate-binding protein from Thermomonospora curvata
To be Published
|
|
4MZH
 
 | |
4MOZ
 
 | |
4NTR
 
 | |
6J3O
 
 | | Crystal structure of the human PCAF bromodomain in complex with compound 12 | | Descriptor: | 3-methyl-2-[[(3~{R})-1-methylpiperidin-3-yl]amino]-5~{H}-pyrrolo[3,2-d]pyrimidin-4-one, Histone acetyltransferase KAT2B | | Authors: | Huang, L.Y, Li, H, Li, L.L, Niu, L, Seupel, R, Wu, C.Y, Li, G.B, Yu, Y.M, Brennan, P.E, Yang, S.Y. | | Deposit date: | 2019-01-05 | | Release date: | 2019-05-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Discovery of Pyrrolo[3,2- d]pyrimidin-4-one Derivatives as a New Class of Potent and Cell-Active Inhibitors of P300/CBP-Associated Factor Bromodomain.
J.Med.Chem., 62, 2019
|
|
4NW8
 
 | |
2AG8
 
 | | NADP complex of Pyrroline-5-carboxylate reductase from Neisseria meningitidis | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, pyrroline-5-carboxylate reductase | | Authors: | Chang, C, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-07-26 | | Release date: | 2005-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Delta(1)-Pyrroline-5-carboxylate Reductase from Human Pathogens Neisseria meningitides and Streptococcus pyogenes.
J.Mol.Biol., 354, 2005
|
|
4HES
 
 | | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula. | | Descriptor: | Beta-lactamase class A-like protein, FORMIC ACID, GLYCEROL, ... | | Authors: | Cuff, M.E, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-10-04 | | Release date: | 2012-10-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Beta-Lactamase Class A-like Protein from Veillonella parvula.
TO BE PUBLISHED
|
|
4O5A
 
 | | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI family transcription regulator, SULFATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-12-19 | | Release date: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | The crystal structure of a LacI family transcriptional regulator from Bifidobacterium animalis subsp. lactis DSM 10140.
To be Published
|
|
4LS0
 
 | | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DH01B0033 | | Descriptor: | 2-{(E)-[2-(4-phenyl-1,3-thiazol-2-yl)hydrazinylidene]methyl}benzaldehyde, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Zhu, L, Li, H, Ren, X, Zhu, J. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of human dihydroorotate dehydrogenase (DHODH) with DHO1B0033
To be Published
|
|
3RPD
 
 | | The structure of a B12-independent methionine synthase from Shewanella sp. W3-18-1 in complex with Selenomethionine. | | Descriptor: | GLYCEROL, Methionine synthase (B12-independent), SELENOMETHIONINE, ... | | Authors: | Cuff, M.E, Li, H, Hatzos-Skintges, C, Tesar, C, Bearden, J, Clancy, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-31 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of a B12-independent methionine synthase from Shewanella sp. W3-18-1 in complex with Selenomethionine.
TO BE PUBLISHED
|
|
4OG2
 
 | | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with leucine | | Descriptor: | Amino acid/amide ABC transporter substrate-binding protein, HAAT family, CHLORIDE ION, ... | | Authors: | Tan, K, Li, H, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | The crystal structure of a solute-binding protein (N280D mutant) from Anabaena variabilis ATCC 29413 in complex with leucine
To be Published
|
|
