1AGW
 
 | |
2WY2
 
 | | NMR structure of the IIAchitobiose-IIBchitobiose phosphoryl transition state complex of the N,N'-diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT, PHOSPHITE ION | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
4N3Z
 
 | |
4N3X
 
 | |
4N3Y
 
 | |
4Q9U
 
 | | Crystal structure of the Rab5, Rabex-5delta and Rabaptin-5C21 complex | | Descriptor: | Rab GTPase-binding effector protein 1, Rab5 GDP/GTP exchange factor, Ras-related protein Rab-5A | | Authors: | Zhang, Z, Zhang, T, Ding, J. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.618 Å) | | Cite: | Molecular mechanism for Rabex-5 GEF activation by Rabaptin-5
Elife, 3, 2014
|
|
2WWV
 
 | | NMR structure of the IIAchitobiose-IIBchitobiose complex of the N,N'- diacetylchitoboise brance of the E. coli phosphotransferase system. | | Descriptor: | N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIA COMPONENT, N,N'-DIACETYLCHITOBIOSE-SPECIFIC PHOSPHOTRANSFERASE ENZYME IIB COMPONENT | | Authors: | Sang, Y.S, Cai, M, Clore, G.M. | | Deposit date: | 2009-10-29 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Iiachitobose-Iibchitobiose Complex of the N,N'-Diacetylchitobiose Branch of the Escherichia Coli Phosphotransfer System
J.Biol.Chem., 285, 2010
|
|
6A58
 
 | | Structure of histone demethylase REF6 | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
6A59
 
 | | Structure of histone demethylase REF6 at 1.8A | | Descriptor: | Lysine-specific demethylase REF6, ZINC ION | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
6A57
 
 | | Structure of histone demethylase REF6 complexed with DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*TP*CP*TP*CP*TP*GP*TP*TP*TP*TP*GP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*AP*AP*AP*CP*AP*GP*AP*GP*AP*AP*A)-3'), GLYCEROL, ... | | Authors: | Tian, Z, Chen, Z. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of REF6 and its complex with DNA reveal diverse recognition mechanisms.
Cell Discov, 6, 2020
|
|
4NX4
 
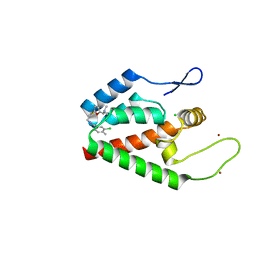 | | Re-refinement of CAP-1 HIV-CA complex | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-{2-[({5-[(dimethylamino)methyl]-2-furyl}methyl)thio]ethyl}urea, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Lang, P.T, Holton, J.M, Fraser, J.S, Alber, T. | | Deposit date: | 2013-12-08 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein structural ensembles are revealed by redefining X-ray electron density noise.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6JG8
 
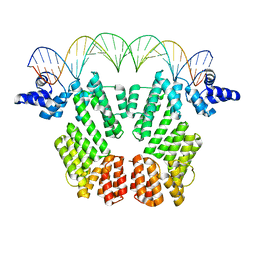 | | Crystal structure of AimR in complex with DNA | | Descriptor: | AimR transcriptional regulator, DNA (31-MER) | | Authors: | Guan, Z.Y, Pei, K, Zou, T.T. | | Deposit date: | 2019-02-13 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural insights into DNA recognition by AimR of the arbitrium communication system in the SPbeta phage.
Cell Discov, 5, 2019
|
|
6JG9
 
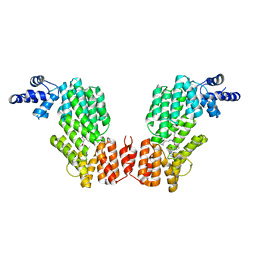 | |
6JG5
 
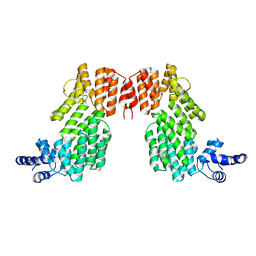 | | Crystal structure of AimR | | Descriptor: | AimR transcriptional regulator | | Authors: | Guan, Z.Y, Pei, K, Zou, T.T. | | Deposit date: | 2019-02-13 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Structural insights into DNA recognition by AimR of the arbitrium communication system in the SPbeta phage.
Cell Discov, 5, 2019
|
|
6K0R
 
 | | Ruvbl1-Ruvbl2 with truncated domain II in complex with phosphorylated Cordycepin | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhang, W, Chen, L, Li, W, Ju, D, Huang, N, Zhang, E. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Chemical perturbations reveal that RUVBL2 regulates the circadian phase in mammals.
Sci Transl Med, 12, 2020
|
|
3K4T
 
 | |
2MM3
 
 | | Solution NMR structure of the ternary complex of human ileal bile acid-binding protein with glycocholate and glycochenodeoxycholate | | Descriptor: | GLYCOCHENODEOXYCHOLIC ACID, GLYCOCHOLIC ACID, Gastrotropin | | Authors: | Horvath, G, Egyed, O, Bencsura, A, Simon, A, Tochtrop, G.P, DeKoster, G.T, Covey, D.F, Cistola, D.P, Toke, O. | | Deposit date: | 2014-03-07 | | Release date: | 2014-05-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of ligand binding in the ternary complex of human ileal bile acid binding protein with glycocholate and glycochenodeoxycholate obtained from solution NMR
FEBS J., 283, 2016
|
|
3F6N
 
 | |
3WBA
 
 | |
3WBE
 
 | |
1QLR
 
 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF A HUMAN MONOCLONAL IgM COLD AGGLUTININ | | Descriptor: | IGM FAB REGION IV-J(H4)-C (KAU COLD AGGLUTININ), IGM KAPPA CHAIN V-III (KAU COLD AGGLUTININ), alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Carvalho, J.G, Cauerhff, A, Goldbaum, F, Leoni, J, Polikarpov, I. | | Deposit date: | 1999-09-11 | | Release date: | 2000-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Three-dimensional structure of the Fab from a human IgM cold agglutinin.
J Immunol., 165, 2000
|
|
