3OWO
 
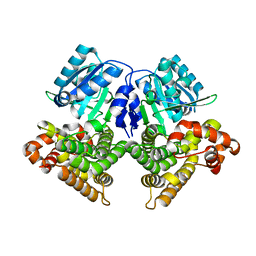 | | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD cofactor | | Descriptor: | Alcohol dehydrogenase 2, FE (II) ION | | Authors: | Moon, J.H, Lee, H.J, Song, J.M, Park, S.Y, Park, M.Y, Park, H.M, Sun, J, Park, J.H, Kim, J.S. | | Deposit date: | 2010-09-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor
J.Mol.Biol., 407, 2011
|
|
3PST
 
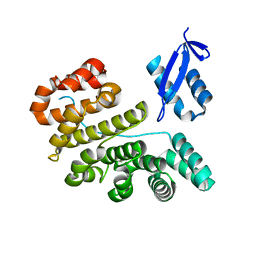 | |
3PSP
 
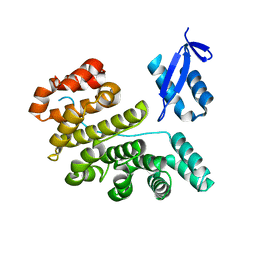 | |
5U8T
 
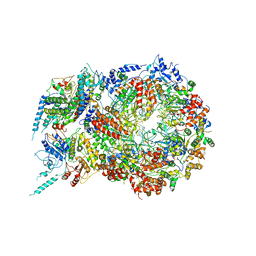 | | Structure of Eukaryotic CMG Helicase at a Replication Fork and Implications | | Descriptor: | Cell division control protein 45, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication complex GINS protein PSF1, ... | | Authors: | Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, Li, H, O'Donnell, M.E. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UJM
 
 | | Structure of the active form of human Origin Recognition Complex and its ATPase motor module | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Tocilj, A, On, K, Yuan, Z, Sun, J, Elkayam, E, Li, H, Stillman, B, Joshua-Tor, L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
5U8S
 
 | | Structure of eukaryotic CMG helicase at a replication fork | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (26-MER), ... | | Authors: | Li, H, Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, O'Donnell, M.E. | | Deposit date: | 2016-12-14 | | Release date: | 2017-01-25 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (6.101 Å) | | Cite: | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7WHP
 
 | | Pentameric turret of Bombyx mori cytoplasmic polyhedrosis virus after spike detaches. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, S-ADENOSYLMETHIONINE, Structural protein VP3 | | Authors: | Zhang, Y, Cui, Y, Sun, J, Zhou, Z.H. | | Deposit date: | 2021-12-31 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Multiple conformations of trimeric spikes visualized on a non-enveloped virus.
Nat Commun, 13, 2022
|
|
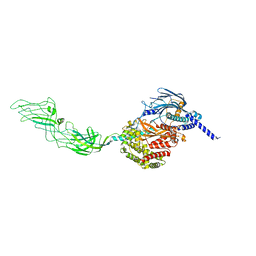
7WHM
 
 | |
7WHN
 
 | |
7W7H
 
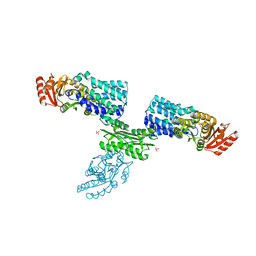 | | S Suis FakA-FakB2 complex structure | | Descriptor: | OLEIC ACID, Predicted kinase related to dihydroxyacetone kinase, SULFATE ION, ... | | Authors: | Shi, Y, Zang, N, Lou, N, Xu, Y, Sun, J, Huang, M, Zhang, H, Lu, H, Zhou, C, Feng, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism for streptococcal fatty acid kinase (Fak) system dedicated to host fatty acid scavenging.
Sci Adv, 8, 2022
|
|
7CQ2
 
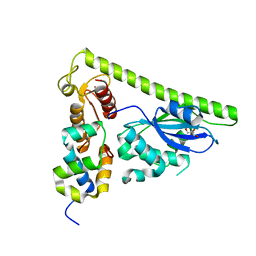 | | Crystal structure of Slx1-Slx4 | | Descriptor: | GLYCEROL, SLX4 isoform 1, Structure-specific endonuclease subunit SLX1, ... | | Authors: | Xu, X, Wang, M, Sun, J, Li, G, Yang, N, Xu, R.M. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure specific DNA recognition by the SLX1-SLX4 endonuclease complex.
Nucleic Acids Res., 49, 2021
|
|
7CQ3
 
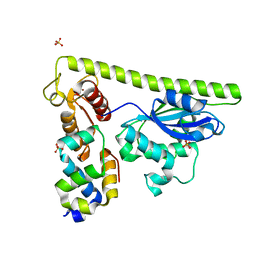 | | Crystal structure of Slx1-Slx4 | | Descriptor: | SLX4 isoform 1, SULFATE ION, Structure-specific endonuclease subunit SLX1, ... | | Authors: | Xu, X, Wang, M, Sun, J, Li, G, Yang, N, Xu, R.M. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Structure specific DNA recognition by the SLX1-SLX4 endonuclease complex.
Nucleic Acids Res., 49, 2021
|
|
7CQ4
 
 | | Crystal structure of Slx1-Slx4 in complex with 5'flap DNA | | Descriptor: | DNA (27-MER), DNA (5'-D(*AP*GP*GP*AP*CP*AP*TP*CP*TP*TP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*TP*AP*CP*AP*AP*CP*AP*GP*AP*T)-3'), ... | | Authors: | Xu, X, Wang, M, Sun, J, Li, G, Yang, N, Xu, R.M. | | Deposit date: | 2020-08-08 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.294 Å) | | Cite: | Structure specific DNA recognition by the SLX1-SLX4 endonuclease complex.
Nucleic Acids Res., 49, 2021
|
|
7BW8
 
 | | Cryo-EM Structure for the Insulin Binding Region in the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BWA
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 2 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
6V97
 
 | | Kindlin-3 double deletion mutant short form | | Descriptor: | Fermitin family homolog 3 | | Authors: | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | Deposit date: | 2019-12-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.381 Å) | | Cite: | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
6V9G
 
 | | Kindlin-3 double deletion mutant long form | | Descriptor: | Fermitin family homolog 3 | | Authors: | Xu, Z, Zhang, T.L, Xu, Z, Sun, J.J, Ding, J.P, Ma, Y.Q. | | Deposit date: | 2019-12-13 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure basis of the FERM domain of kindlin-3 in supporting integrin alpha IIb beta 3 activation in platelets.
Blood Adv, 4, 2020
|
|
7BW6
 
 | | Varicella-zoster virus capsid | | Descriptor: | Major capsid protein, Small capsomere-interacting protein, Triplex capsid protein 1, ... | | Authors: | Wang, P.Y, Qi, J.X, Liu, C.C, Sun, J.Q. | | Deposit date: | 2020-04-13 | | Release date: | 2020-09-23 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the varicella-zoster virus A-capsid.
Nat Commun, 11, 2020
|
|
7RKI
 
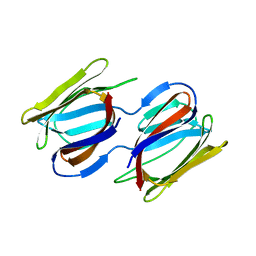 | | Griffithsin-S10Y/S42Y/S88Y | | Descriptor: | Griffithsin, alpha-D-mannopyranose | | Authors: | Sun, J.D, Zhao, G.X. | | Deposit date: | 2021-07-22 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7XUR
 
 | |
5FI7
 
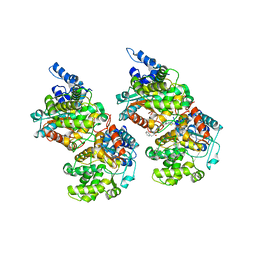 | | Crystal structure of human GAC in complex with inhibitor UPGL_00015: 2-phenyl-~{N}-[5-[(3~{S})-3-[[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]oxy]pyrrolidin-1-yl]-1,3,4-thiadiazol-2-yl]ethanamide | | Descriptor: | 2-phenyl-~{N}-[5-[(3~{S})-3-[[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]oxy]pyrrolidin-1-yl]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R. | | Deposit date: | 2015-12-22 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and evaluation of novel glutaminase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5FI2
 
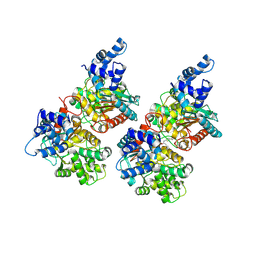 | | Crystal structure of human GAC in complex with inhibitor UPGL_00009: 2-phenyl-~{N}-[5-[[(3~{R})-1-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol- 2-yl]pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide | | Descriptor: | 2-phenyl-~{N}-[5-[[(3~{R})-1-[5-(2-phenylethanoylamino)-1,3,4-thiadiazol-2-yl]pyrrolidin-3-yl]amino]-1,3,4-thiadiazol-2-yl]ethanamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R. | | Deposit date: | 2015-12-22 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and evaluation of novel glutaminase inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
8ID3
 
 | | Cryo-EM structure of the 9-hydroxystearic acid bound GPR120-Gi complex | | Descriptor: | 9-Hydroxyoctadecanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID9
 
 | | Cryo-EM structure of the eicosapentaenoic acid bound GPR120-Gi complex | | Descriptor: | 5,8,11,14,17-EICOSAPENTAENOIC ACID, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
8ID8
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Gi complex | | Descriptor: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | Deposit date: | 2023-02-12 | | Release date: | 2023-03-15 | | Last modified: | 2023-05-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
