5H2O
 
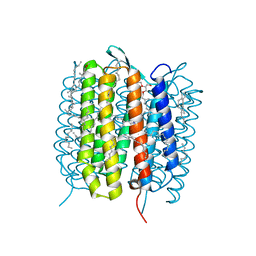 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 250 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2M
 
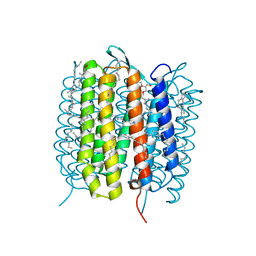 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 13.8 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2J
 
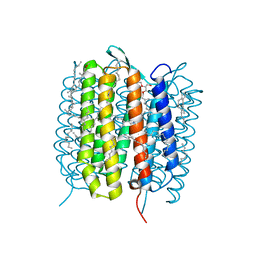 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 290 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2P
 
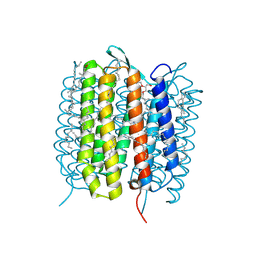 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 657 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2H
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 40 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
8J2V
 
 | |
8J2U
 
 | |
8J2Z
 
 | | Glucosyl transferase | | Descriptor: | Glycosyltransferase | | Authors: | Arold, S.T, Hameed, U.F.S. | | Deposit date: | 2023-04-15 | | Release date: | 2024-04-17 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.783 Å) | | Cite: | beta-Carotene alleviates substrate inhibition caused by asymmetric cooperativity.
Nat Commun, 16, 2025
|
|
8J31
 
 | |
5H2K
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
5H2I
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 110 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-10-15 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
8YA2
 
 | | Structure of the SecA-SecY complex with the substrate FtsQ-LacY(+20C) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division protein FtsQ,Lactose permease, ... | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
8YA0
 
 | | Structure of the SecA-SecY complex with the substrate FtsQ-LacY(+7C) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cell division protein FtsQ,Lactose permease, ... | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
8YA3
 
 | | Structure of the SecA-SecY complex with the substrate FtsQ-LacY(+7C) treated with DTT | | Descriptor: | Cell division protein FtsQ,Lactose permease, Protein translocase subunit SecE, Protein translocase subunit SecY | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
8YAS
 
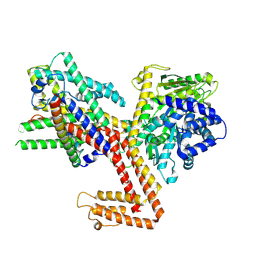 | | Structure of the SecA-SecY complex with the substrate HmBRI-7TM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-10 | | Release date: | 2025-02-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
8Y9Z
 
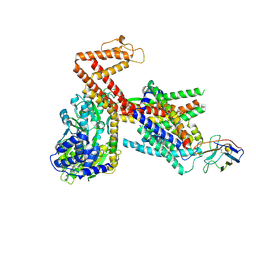 | | Structure of the SecA-SecY complex with the substrate HmBRI-3TM | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Bacteriorhodopsin-I, ... | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
8Y9Y
 
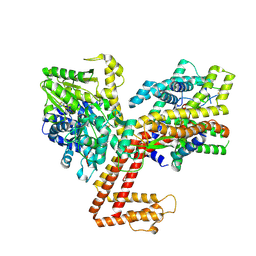 | | Structure of the SecA-SecY complex with the substrate FtsQ-LacY(+1C) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Ou, X, Ma, C, Sun, D, Xu, J, Wu, X, Gao, N, Li, L. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | SecY translocon chaperones protein folding during membrane protein insertion.
Cell, 188, 2025
|
|
9IIO
 
 | |
6JPF
 
 | |
5Z1F
 
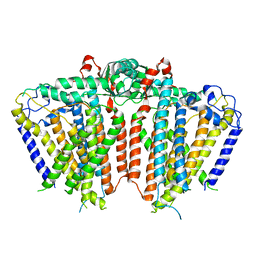 | |
5GZE
 
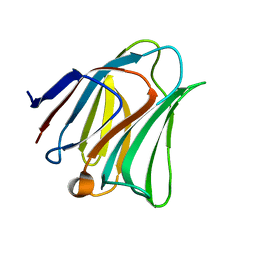 | | Galectin-8 N-terminal domain carbohydrate recognition domain | | Descriptor: | Galectin-8, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J.Y, Si, Y.L. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Crystallization of Galectin-8 Linker Reveals Intricate Relationship between the N-terminal Tail and the Linker.
Int J Mol Sci, 17, 2016
|
|
8AHH
 
 | |
8AHG
 
 | |
8AHI
 
 | |
5H18
 
 | | Crystal structure of catalytic domain of UGGT (UDP-glucose-bound form) from Thermomyces dupontii | | Descriptor: | CALCIUM ION, GLYCEROL, UGGT, ... | | Authors: | Satoh, T, Zhu, T, Toshimori, T, Kamikubo, H, Uchihashi, T, Kato, K. | | Deposit date: | 2016-10-08 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Visualisation of a flexible modular structure of the ER folding-sensor enzyme UGGT.
Sci Rep, 7, 2017
|
|
