5AWO
 
 | | Arthrobacter globiformis T6 isomalto-dextranse | | Descriptor: | ACETATE ION, Isomaltodextranase, PHOSPHATE ION | | Authors: | Tonozuka, T. | | Deposit date: | 2015-07-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal Structure and Mutational Analysis of Isomalto-dextranase, a Member of Glycoside Hydrolase Family 27
J.Biol.Chem., 290, 2015
|
|
5AWP
 
 | |
5AWQ
 
 | | Arthrobacter globiformis T6 isomalto-dextranse complexed with panose | | Descriptor: | Isomaltodextranase, PHOSPHATE ION, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Tonozuka, T. | | Deposit date: | 2015-07-08 | | Release date: | 2015-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal Structure and Mutational Analysis of Isomalto-dextranase, a Member of Glycoside Hydrolase Family 27
J.Biol.Chem., 290, 2015
|
|
4ZTD
 
 | | Crystal Structure of Human PCNA in complex with a TRAIP peptide | | Descriptor: | ALA-GLY-ALA-GLY-ALA, ALA-PHE-GLN-ALA-LYS-LEU-ASP-THR-PHE-LEU-TRP-SER, Proliferating cell nuclear antigen | | Authors: | Montoya, G, Mortuza, G.B, Blanco, F.J, Ibanez de Opakua, A. | | Deposit date: | 2015-05-14 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress.
J.Cell Biol., 212, 2016
|
|
8DPR
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with inhibitor TKB-248 | | Descriptor: | 2,2,2-trifluoro-N-{(2S)-1-[(1R,2S,5S)-2-({(2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamothioyl)-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-3-yl]-3,3-dimethyl-1-oxobutan-2-yl}acetamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, ... | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-16 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8DOX
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Bulut, H, Hayashi, H, Tsuji, K, Kuwata, N, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2022-07-14 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Identification of SARS-CoV-2 M pro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2.
Nat Commun, 14, 2023
|
|
6L9W
 
 | | Crystal structure of mouse TIFA (T9E/C36S mutant) | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
6L9V
 
 | | Crystal structure of mouse TIFA (T9D/C36S mutant) | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
6L9U
 
 | | Crystal structure of mouse TIFA | | Descriptor: | TRAF-interacting protein with FHA domain-containing protein A | | Authors: | Nakamura, T, Yamagata, Y. | | Deposit date: | 2019-11-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural analysis of TIFA: Insight into TIFA-dependent signal transduction in innate immunity.
Sci Rep, 10, 2020
|
|
4W4O
 
 | | High-resolution crystal structure of Fc bound to its human receptor Fc-gamma-RI | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Kiyoshi, M, Tsumoto, K. | | Deposit date: | 2014-08-15 | | Release date: | 2015-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for binding of human IgG1 to its high-affinity human receptor Fc gamma RI
Nat Commun, 6, 2015
|
|
5K9J
 
 | | Crystal structure of multidonor HV6-1-class broadly neutralizing Influenza A antibody 56.a.09 isolated following H5 immunization. | | Descriptor: | 56.a.09 heavy chain, 56.a.09 light chain, POLYETHYLENE GLYCOL (N=34) | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-31 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
5K9K
 
 | | Crystal structure of multidonor HV6-1-class broadly neutralizing Influenza A antibody 56.a.09 in complex with Hemagglutinin Hong Kong 1968. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 56.a.09 Heavy chain, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-05-31 | | Release date: | 2016-12-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
5KAN
 
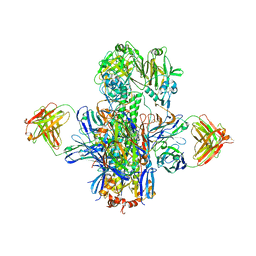 | | Crystal structure of multidonor HV1-18-class broadly neutralizing Influenza A antibody 16.g.07 in complex with A/Hong Kong/1-4-MA21-1/1968 (H3N2) Hemagglutinin | | Descriptor: | 16.g.07 Heavy chain, 16.g.07 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.785 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell(Cambridge,Mass.), 166, 2016
|
|
5KAQ
 
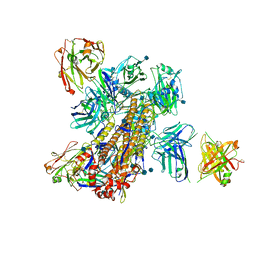 | | Crystal structure of broadly neutralizing Influenza A antibody 31.a.83 in complex with Hemagglutinin Hong Kong 1968. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 31.A.83 FAB HEAVY CHAIN, ANTIBODY 31.A.83 FAB LIGHT CHAIN, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.514 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
5K9O
 
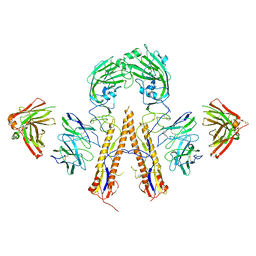 | | Crystal structure of multidonor HV1-18+HD3-9 class broadly neutralizing Influenza A antibody 31.b.09 in complex with Hemagglutinin H1 A/California/04/2009 | | Descriptor: | 31.b.09 Heavy Fv, 31.b.09 Light Fv, Hemagglutinin, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.387 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell, 166, 2016
|
|
5K9Q
 
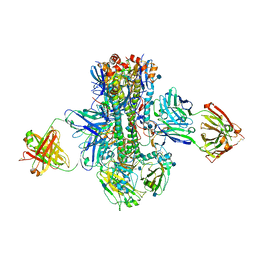 | | Crystal structure of multidonor HV1-18-class broadly neutralizing Influenza A antibody 16.a.26 in complex with A/Hong Kong/1-4-MA21-1/1968 (H3N2) Hemagglutinin | | Descriptor: | 16.a.26 Heavy chain, 16.a.26 Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Joyce, M.G, Thomas, P.V, Wheatley, A.K, McDermott, A.B, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Vaccine-Induced Antibodies that Neutralize Group 1 and Group 2 Influenza A Viruses.
Cell(Cambridge,Mass.), 166, 2016
|
|
8UH8
 
 | | Crystal structure of SARS-CoV-2 main protease E166V (Apo structure) | | Descriptor: | ORF1a polyprotein | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
8UH5
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-06 | | Release date: | 2023-12-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
8UH9
 
 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-272 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(5-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.067 Å) | | Cite: | TKB272, an Orally Available SARS-CoV-2-Mpro Inhibitor Containing 5-Fluorobenzothiazole, Potently Blocks SARS-CoV-2 Replication without Ritonavir
To Be Published
|
|
5J37
 
 | | Crystal structure of 60-mer BFDV Capsid Protein in complex with single stranded DNA | | Descriptor: | Beak and feather disease virus capsid protein, PHOSPHATE ION, single stranded DNA | | Authors: | Sarker, S, Raidal, S, Aragao, D, Forwood, J.K. | | Deposit date: | 2016-03-30 | | Release date: | 2016-05-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the assembly and regulation of distinct viral capsid complexes.
Nat Commun, 7, 2016
|
|
8ILL
 
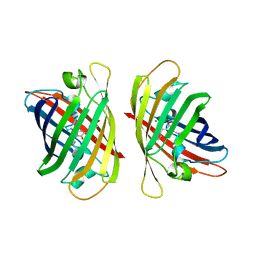 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH5.6 | | Descriptor: | CHLORIDE ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose, green fluorescent protein | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
8ILK
 
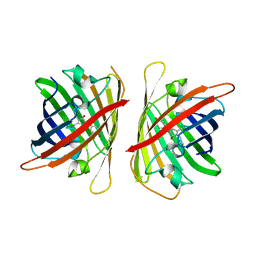 | | Crystal structure of a highly photostable and bright green fluorescent protein at pH8.5 | | Descriptor: | CHLORIDE ION, Green FLUORESCENT PROTEIN | | Authors: | Ago, H, Ando, R, Hirano, M, Shimozono, S, Miyawaki, A, Yamamoto, M. | | Deposit date: | 2023-03-03 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | StayGold variants for molecular fusion and membrane-targeting applications.
Nat.Methods, 21, 2024
|
|
9JR3
 
 | | Cryo-EM structure of PTH-PTH1R-Gq (tilted state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sano, F.K, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-09-29 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Insights into G-protein coupling preference from cryo-EM structures of Gq-bound PTH1R
To Be Published
|
|
9JR2
 
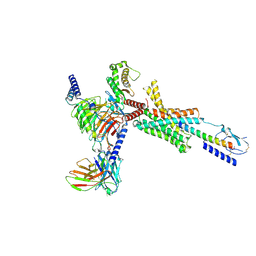 | | Cryo-EM structure of PTH-PTH1R-Gq (upright state) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(q) subunit alpha-1 (miniGq), ... | | Authors: | Sano, F.K, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-09-29 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Insights into G-protein coupling preference from cryo-EM structures of Gq-bound PTH1R
To Be Published
|
|
3PCV
 
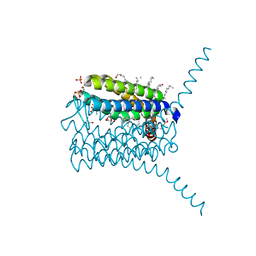 | | Crystal structure analysis of human leukotriene C4 synthase at 1.9 angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, DODECYL-BETA-D-MALTOSIDE, GLUTATHIONE, ... | | Authors: | Saino, H, Ago, H, Miyano, M. | | Deposit date: | 2010-10-22 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The catalytic architecture of leukotriene C4 synthase with two arginine residues
J.Biol.Chem., 286, 2011
|
|
