8BBS
 
 | | Structure of AKR1C3 in complex with a bile acid fused tetrazole inhibitor | | Descriptor: | (4~{R})-4-[(1~{R},2~{S},5~{R},6~{R},13~{S},14~{S},17~{R},19~{R})-6,14-dimethyl-17-oxidanyl-7,8,9,10-tetrazapentacyclo[11.8.0.0^{2,6}.0^{7,11}.0^{14,19}]henicosa-8,10-dien-5-yl]pentanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Petri, E.T, Skerlova, J, Marinovic, M, Brynda, J, Kugler, M, Skoric, D, Bekic, S, Celic, A.S, Rezacova, P. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray structure of human aldo-keto reductase 1C3 in complex with a bile acid fused tetrazole inhibitor: experimental validation, molecular docking and structural analysis.
Rsc Med Chem, 14, 2023
|
|
4K2A
 
 | | Crystal structure of haloalkane dehalogenase DbeA from Bradyrhizobium elkani USDA94 | | Descriptor: | ACETATE ION, CHLORIDE ION, Haloalkane dehalogenase | | Authors: | Prudnikova, T, Chaloupkova, R, Rezacova, P, Mozga, T, Koudelakova, T, Sato, Y, Kuty, M, Nagata, Y, Damborsky, J, Kuta Smatanova, I, Structure 2 Function Project (S2F) | | Deposit date: | 2013-04-08 | | Release date: | 2014-06-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analysis of a novel haloalkane dehalogenase with two halide-binding sites.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4LL3
 
 | | Structure of wild-type HIV protease in complex with darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, Protease | | Authors: | Grantz Saskova, K, Rezacova, P, Brynda, J, Kozisek, M, Konvalinka, J. | | Deposit date: | 2013-07-09 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thermodynamic and structural analysis of HIV protease resistance to darunavir - analysis of heavily mutated patient-derived HIV-1 proteases.
Febs J., 281, 2014
|
|
4YBF
 
 | | Aspartic Proteinase Sapp2 Secreted from Candida Parapsilosis at 1.25 A Resolution | | Descriptor: | Candidapepsin-2, DI(HYDROXYETHYL)ETHER | | Authors: | Dostal, J, Hruskova-Heidingsfeldova, O, Rezacova, P, Brynda, J, Mareckova, L, Pichova, I. | | Deposit date: | 2015-02-18 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Atomic resolution crystal structure of Sapp2p, a secreted aspartic protease from Candida parapsilosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Y9W
 
 | | Aspartic Proteinase Sapp2 Secreted from Candida Parapsilosis at 0.82 A Resolution. | | Descriptor: | Aspartic acid endopeptidase Sapp2, PEPTIDE | | Authors: | Dostal, J, Brynda, J, Hruskova-Heidingsfeldova, O, Rezacova, P, Mareckova, L, Pichova, I. | | Deposit date: | 2015-02-17 | | Release date: | 2016-11-30 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (0.83 Å) | | Cite: | Atomic resolution crystal structure of Sapp2p, a secreted aspartic protease from Candida parapsilosis.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8A2K
 
 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-[(2-naphthyloxy)methyl]phenyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-[4-(naphthalen-1-yloxymethyl)phenyl]pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-3~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2J
 
 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-biphenylyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-(4-phenylphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2I
 
 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-(2-naphthyl)phenyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-(4-naphthalen-2-ylphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-3~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
7ACK
 
 | | CDK2/cyclin A2 in complex with an imidazo[1,2-c]pyrimidin-5-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 8-cyclohexyl-6~{H}-imidazo[1,2-c]pyrimidin-5-one, Cyclin-A2, ... | | Authors: | Skerlova, J, Pachl, P, Rezacova, P. | | Deposit date: | 2020-09-11 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Imidazo[1,2-c]pyrimidin-5(6H)-one inhibitors of CDK2: Synthesis, kinase inhibition and co-crystal structure.
Eur.J.Med.Chem., 216, 2021
|
|
5OAR
 
 | | Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, ... | | Authors: | Skerlova, J, Rezacova, P, Brynda, J, Pachl, P, Otwinowski, Z, Vanek, O. | | Deposit date: | 2017-06-23 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of native beta-N-acetylhexosaminidase isolated from Aspergillus oryzae sheds light onto its substrate specificity, high stability, and regulation by propeptide.
FEBS J., 285, 2018
|
|
5OPL
 
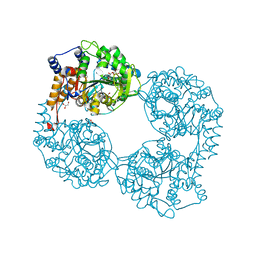 | | Crystal structure of K25E cN-II mutant | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, MAGNESIUM ION | | Authors: | Kugler, M, Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPM
 
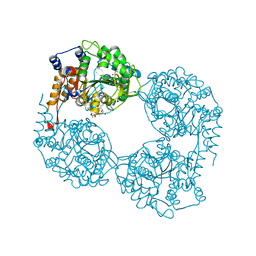 | | Crystal structure of D52N/R238W cN-II mutant bound to dATP and free phosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPK
 
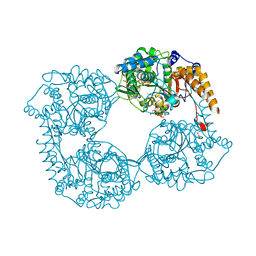 | | Crystal structure of D52N/R367Q cN-II mutant bound to dATP and free phosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPP
 
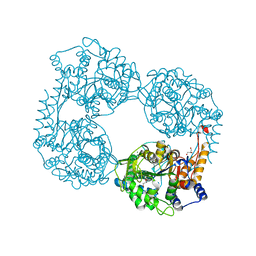 | | Crystal structure of S408R cN-II mutant | | Descriptor: | BROMIDE ION, Cytosolic purine 5'-nucleotidase, GLYCEROL, ... | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPN
 
 | | Crystal structure of R39Q cN-II mutant | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5OPO
 
 | | Crystal structure of R238G cN-II mutant | | Descriptor: | Cytosolic purine 5'-nucleotidase, GLYCEROL, MAGNESIUM ION | | Authors: | Hnizda, A, Pachl, P, Rezacova, P. | | Deposit date: | 2017-08-10 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Relapsed acute lymphoblastic leukemia-specific mutations in NT5C2 cluster into hotspots driving intersubunit stimulation.
Leukemia, 32, 2018
|
|
5NXM
 
 | | Carbonic Anhydrase II Inhibitor RA1 | | Descriptor: | 8-methoxy-2-oxidanylidene-~{N}-(4-sulfamoylphenyl)chromene-3-carboxamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-10 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXI
 
 | | Carbonic Anhydrase II Inhibitor RA2 | | Descriptor: | 4-[(4-bromophenyl)sulfonylamino]benzenesulfonamide, CITRIC ACID, Carbonic anhydrase 2, ... | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXO
 
 | | Carbonic Anhydrase II Inhibitor RA6 | | Descriptor: | 4-[(4-nitrophenyl)methyl]benzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXV
 
 | | Carbonic Anhydrase II Inhibitor RA8 | | Descriptor: | 2-(4-chloranyl-5-methyl-3-nitro-pyrazol-1-yl)-~{N}-(4-sulfamoylphenyl)ethanamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NY3
 
 | | Carbonic Anhydrase II Inhibitor RA11 | | Descriptor: | 1-(4-chlorophenyl)-3-[2-(4-sulfamoylphenyl)ethyl]urea, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NYA
 
 | | Carbonic Anhydrase II Inhibitor RA13 | | Descriptor: | Carbonic anhydrase 2, GLUTARIC ACID, ZINC ION, ... | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NY1
 
 | | Carbonic Anhydrase II Inhibitor RA10 | | Descriptor: | 3,6,7-trimethyl-~{N}-[(4-sulfamoylphenyl)methyl]-1-benzofuran-2-carboxamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NY6
 
 | | Carbonic Anhydrase II Inhibitor RA12 | | Descriptor: | 4-chloranyl-~{N}-[(1~{R})-1-(2-hydroxyphenyl)ethyl]-3-sulfamoyl-benzamide, 4-chloranyl-~{N}-[(1~{S})-1-(2-hydroxyphenyl)ethyl]-3-sulfamoyl-benzamide, Carbonic anhydrase 2, ... | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-11 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
5NXG
 
 | | Carbonic Anhydrase II Inhibitor RA1 | | Descriptor: | 2-chloranyl-4-nitro-~{N}-(4-sulfamoylphenyl)benzamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Brynda, J, Rezacova, P, Horejsi, M, Fanfrlik, J. | | Deposit date: | 2017-05-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ranking Power of the SQM/COSMO Scoring Function on Carbonic Anhydrase II-Inhibitor Complexes.
Chemphyschem, 19, 2018
|
|
