4LAT
 
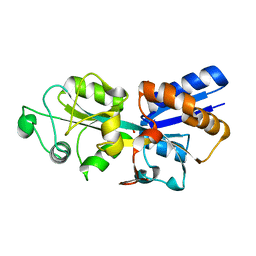 | | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A in complex with phosphate | | Descriptor: | ISOPROPYL ALCOHOL, PHOSPHATE ION, Phosphate-binding protein PstS 1 | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of phosphate ABC transporter, periplasmic phosphate-binding protein PstS 1 (PBP1) from Streptococcus pneumoniae Canada MDR_19A in complex with phosphate
To be Published
|
|
3TSD
 
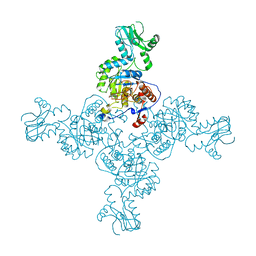 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames complexed with XMP | | Descriptor: | D(-)-TARTARIC ACID, Inosine-5'-monophosphate dehydrogenase, SULFATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3TSB
 
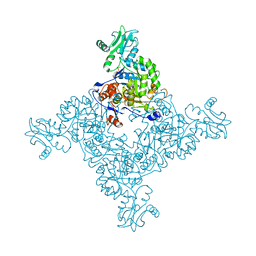 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames | | Descriptor: | Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3TV9
 
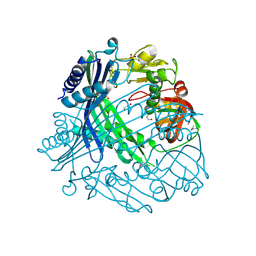 | | Crystal Structure of Putative Peptide Maturation Protein from Shigella flexneri | | Descriptor: | GLYCEROL, Putative peptide maturation protein, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal Structure of Putative Peptide Maturation Protein from
To be Published
|
|
3H2Y
 
 | | Crystal structure of YqeH GTPase from Bacillus anthracis with dGDP bound | | Descriptor: | 2'-DEOXYGUANOSINE-5'-DIPHOSPHATE, GTPase family protein | | Authors: | Brunzelle, J.S, Anderson, S.M, Xu, X, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of YqeH GTPase from Bacillus anthracis with dGDP Bound
To be Published
|
|
3OOW
 
 | | Octameric structure of the phosphoribosylaminoimidazole carboxylase catalytic subunit from Francisella tularensis subsp. tularensis SCHU S4. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, F.W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Octameric structure of the phosphoribosylaminoimidazole carboxylase catalytic subunit from Francisella tularensis subsp. tularensis SCHU S4.
To be Published
|
|
3SKS
 
 | | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames | | Descriptor: | PHOSPHATE ION, Putative Oligoendopeptidase F, ZINC ION | | Authors: | Wajerowicz, W, Onopriyenko, O, Porebski, P, Domagalski, M, Chruszcz, M, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-23 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a putative oligoendopeptidase F from Bacillus anthracis str. Ames
TO BE PUBLISHED
|
|
3T7Y
 
 | | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis | | Descriptor: | CHLORIDE ION, FORMIC ACID, SODIUM ION, ... | | Authors: | Singer, A.U, Wawrzak, Z, Skarina, T, Saikali, P, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-31 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of an autocleavage-inactive mutant of the cytoplasmic domain of CT091, the YscU homologue of Chlamydia trachomatis
TO BE PUBLISHED
|
|
3SZ3
 
 | | Crystal structure of Tryptophanyl-tRNA synthetase from Vibrio cholerae with an endogenous tryptophan | | Descriptor: | GLYCEROL, TRYPTOPHAN, Tryptophanyl-tRNA synthetase | | Authors: | Cooper, D.R, Kudritska, M, Chruszcz, M, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-18 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of Tryptophanyl-tRNA synthetase from Vibrio cholerae with an endogenous tryptophan
To be Published
|
|
3SR3
 
 | | Crystal structure of the w180a mutant of microcin immunity protein mccf from Bacillus anthracis shows the active site loop in the open conformation. | | Descriptor: | Microcin immunity protein MccF | | Authors: | Nocek, B, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-06 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Structural and functional characterization of microcin C resistance peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
5UC7
 
 | | Crystal structure of BioA / 7,8-diaminopelargonic acid aminotransferase / DAPA synthase from Citrobacter rodentium, PLP complex | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, D(-)-TARTARIC ACID | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Crystal structure of BioA / 7,8-diaminopelargonic acid aminotransferase / DAPA synthase from Citrobacter rodentium, PLP complex
To Be Published
|
|
3V4E
 
 | | Crystal Structure of the galactoside O-acetyltransferase in complex with CoA | | Descriptor: | COENZYME A, DI(HYDROXYETHYL)ETHER, Galactoside O-acetyltransferase, ... | | Authors: | Knapik, A.A, Shumilin, I.A, Luo, H.-B, Chruszcz, M, Zimmerman, M.D, Cymborowski, M, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-14 | | Release date: | 2012-01-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biophysical analysis of the putative acetyltransferase SACOL2570 from methicillin-resistant Staphylococcus aureus.
J.Struct.Funct.Genom., 14, 2013
|
|
3V4Z
 
 | | D-alanine--D-alanine ligase from Yersinia pestis | | Descriptor: | D-alanine--D-alanine ligase, DI(HYDROXYETHYL)ETHER, TRIETHYLENE GLYCOL | | Authors: | Osipiuk, J, Nocek, B, Mulligan, R, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-15 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | D-alanine--D-alanine ligase from Yersinia pestis.
To be Published
|
|
3THA
 
 | | Tryptophan synthase subunit alpha from Campylobacter jejuni. | | Descriptor: | Tryptophan synthase alpha chain | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-18 | | Release date: | 2011-08-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Tryptophan synthase subunit alpha from Campylobacter jejuni.
To be Published
|
|
3TNG
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase from Listeria monocytogenes EGD-e. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Lmo1369 protein, NICKEL (II) ION | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-01 | | Release date: | 2011-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase from Listeria monocytogenes EGD-e.
To be Published
|
|
3TAU
 
 | | Crystal Structure of a Putative Guanylate Monophosphaste Kinase from Listeria monocytogenes EGD-e | | Descriptor: | Guanylate kinase, SODIUM ION, SULFATE ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Kwok, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-04 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of a Putative Guanylate Monophosphaste Kinase from Listeria monocytogenes EGD-e
To be Published
|
|
3UF6
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoD (3'-dephosphocoenzyme A) | | Descriptor: | DEPHOSPHO COENZYME A, Lmo1369 protein | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoD (3'-dephosphocoenzyme A)
To be Published
|
|
5UXD
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium in complex with azithromycin | | Descriptor: | AZITHROMYCIN, CHLORIDE ION, Macrolide 2'-phosphotransferase MphH, ... | | Authors: | Stogios, P.J, Skarina, T, Wawrzak, Z, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5UXA
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphB from Escherichia coli | | Descriptor: | CALCIUM ION, Macrolide 2'-phosphotransferase II | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
5USX
 
 | | Crystal structure of thioredoxin-disulfide reductase from Vibrio vulnificus CMCP6 in complex with NADP and FAD | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Chang, C, Grimshaw, S, Maltseva, N, Mulligan, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-14 | | Release date: | 2017-02-22 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structural genomics of bacterial drug targets: Application of a high-throughput pipeline to solve 58 protein structures from pathogenic and related bacteria.
Microbiol Resour Announc, 2025
|
|
3U80
 
 | | 1.60 Angstrom Resolution Crystal Structure of a 3-Dehydroquinate Dehydratase-like Protein from Bifidobacterium longum | | Descriptor: | 3-dehydroquinate dehydratase, type II | | Authors: | Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-14 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a type II dehydroquinate dehydratase-like protein from Bifidobacterium longum.
J.Struct.Funct.Genom., 14, 2013
|
|
3UFG
 
 | | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glycyl-tRNA synthetase alpha subunit, LEUCINE | | Authors: | Tan, K, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-01 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | The crystal structure of glycyl-tRNA synthetase subunit alpha from Campylobacter jejuni subsp. jejuni NCTC in complex with ATP
To be Published
|
|
2M1Z
 
 | |
5TCG
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate-bound form | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, FORMIC ACID, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
5TCI
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - BRD4592-bound form | | Descriptor: | (2R,3S,4R)-3-(2'-fluoro[1,1'-biphenyl]-4-yl)-4-(hydroxymethyl)azetidine-2-carbonitrile, FORMIC ACID, MALONATE ION, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Wellington, S, Nag, P.P, Fisher, S.L, Schreiber, S.L, Hung, D.T, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
