3IVY
 
 | | Crystal structure of Mycobacterium tuberculosis cytochrome P450 CYP125, p212121 crystal form | | Descriptor: | Cytochrome P450 CYP125, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McLean, K.J, Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.352 Å) | | Cite: | The Structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection.
J.Biol.Chem., 284, 2009
|
|
6T0X
 
 | | Crystal structure of YTHDC1 with fragment 22 (ACA_DC1_001) | | Descriptor: | (3~{S})-~{N}-methylpyrrolidine-3-carboxamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
3WPA
 
 | | Acinetobacter sp. Tol 5 AtaA C-terminal stalk_FL fused to GCN4 adaptors (CstalkFL) | | Descriptor: | CHLORIDE ION, Trimeric autotransporter adhesin | | Authors: | Koiwai, K, Hartmann, M.D, Yoshimoto, S, Nur 'Izzah, N, Suzuki, A, Linke, D, Lupas, A.N, Hori, K. | | Deposit date: | 2014-01-10 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis for Toughness and Flexibility in the C-terminal Passenger Domain of an Acinetobacter Trimeric Autotransporter Adhesin.
J.Biol.Chem., 291, 2016
|
|
6T11
 
 | | Crystal structure of YTHDC1 with fragment 29 (DHU_DC1_218) | | Descriptor: | N-methyl-1H-indole-7-carboxamide, SULFATE ION, YTHDC1 | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
3IWE
 
 | | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD85646 | | Descriptor: | 2,6-dichloro-4-(2-piperazin-1-ylpyridin-4-yl)-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, Glycylpeptide N-tetradecanoyltransferase 1, TETRADECANOYL-COA | | Authors: | Qiu, W, Hutchinson, A, Wernimont, A, Lin, Y.-H, Kania, A, Ravichandran, M, Kozieradzki, I, Cossar, D, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Wyatt, P.G, Ferguson, M.A.J, Frearson, J.A, Brand, S.Y, Robinson, D.A, Bochkarev, A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-02 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of human type-I N-myristoyltransferase with bound myristoyl-CoA and inhibitor DDD85646
To be Published
|
|
3CFT
 
 | |
2MR5
 
 | | Solution NMR Structure of De novo designed Protein, Northeast Structural Genomics Consortium (NESG) Target OR457 | | Descriptor: | De novo designed Protein OR457 | | Authors: | Pulavarti, S, Nivon, L, Maglaqui, M, Janjua, H, Mao, L, Xiao, R, Kornhaber, G, Baker, D, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2014-07-01 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of De novo designed Protein, Northeast Structural Genomics Consortium (NESG) Target OR457
To be Published
|
|
6SE7
 
 | |
3D5N
 
 | | Crystal structure of the Q97W15_SULSO protein from Sulfolobus solfataricus. NESG target SsR125. | | Descriptor: | Q97W15_SULSO | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Lee, D, Foote, R.E, Maglaqui, M, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-16 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Q97W15_SULSO protein from Sulfolobus solfataricus. NESG target SsR125.
To be Published
|
|
2MVO
 
 | | Solution structure of the lantibiotic self-resistance lipoprotein MlbQ from Microbispora ATCC PTA-5024 | | Descriptor: | Putative lipoprotein | | Authors: | Pozzi, R, Schwartz, P, Linke, D, Kulik, A, Nega, M, Wohlleben, W, Stegmann, E, Coles, M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Distinct mechanisms contribute to immunity in the lantibiotic NAI-107 producer strain Microbispora ATCC PTA-5024.
Environ Microbiol, 18, 2016
|
|
3ISH
 
 | |
3WRC
 
 | | Crystal structure of the anaerobic DesB-Protocatechuate (PCA) complex | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, FE (II) ION, Gallate dioxygenase | | Authors: | Sugimoto, K, Senda, M, Kasai, D, Fukuda, M, Masai, E, Senda, T. | | Deposit date: | 2014-02-21 | | Release date: | 2014-04-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Mechanism of Strict Substrate Specificity of an Extradiol Dioxygenase, DesB, Derived from Sphingobium sp. SYK-6
Plos One, 9, 2014
|
|
6SRV
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021144 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2019-09-06 | | Release date: | 2020-06-10 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6SUQ
 
 | | Crystal Structure of TcdB2-TccC3-TEV | | Descriptor: | TcdB2,TccC3,Genome polyprotein | | Authors: | Roderer, D, Schubert, E, Sitsel, O, Raunser, S. | | Deposit date: | 2019-09-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Towards the application of Tc toxins as a universal protein translocation system.
Nat Commun, 10, 2019
|
|
3IW2
 
 | | Crystal structure of Mycobacterium tuberculosis cytochrome P450 CYP125 in complex with econazole | | Descriptor: | 4-[(2R)-2-[(4-chlorobenzyl)oxy]-2-(2,4-dichlorophenyl)ethyl]-1H-imidazole, Cytochrome P450 CYP125, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | McLean, K.J, Levy, C, Munro, A.W, Leys, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Structure of Mycobacterium tuberculosis CYP125: molecular basis for cholesterol binding in a P450 needed for host infection.
J.Biol.Chem., 284, 2009
|
|
6SPC
 
 | | Pseudomonas aeruginosa 30s ribosome from an aminoglycoside resistant clinical isolate | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1E4X
 
 | | crossreactive binding of a circularized peptide to an anti-TGFalpha antibody Fab-fragment | | Descriptor: | CYCLIC PEPTIDE, TAB2 | | Authors: | Hahn, M, Winkler, D, Misselwitz, R, Wessner, H, Welfle, K, Zahn, G, Schneider-Mergener, J, Hoehne, W. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Reactive Binding of Cyclic Peptides to an Anti-Tgf Alpha Antibody Fab Fragment: An X-Ray Structural and Thermodynamic Analysis
J.Mol.Biol., 314, 2001
|
|
6SX5
 
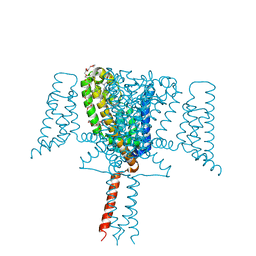 | | Full length Mutant Open-form Sodium Channel NavMs | | Descriptor: | DODECAETHYLENE GLYCOL, HEGA-10, Ion transport protein, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
6SXC
 
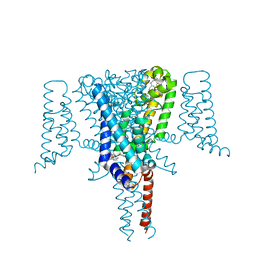 | | Crystal structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with 4-hydroxytamoxifen (2.5 Angstrom resolution) | | Descriptor: | 4-HYDROXYTAMOXIFEN, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2019-09-25 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
3I9A
 
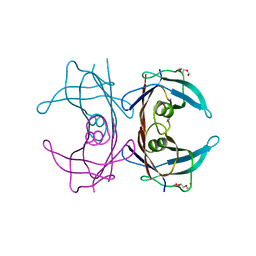 | | Crystal structure of human transthyretin variant A25T - #1 | | Descriptor: | TRIETHYLENE GLYCOL, Transthyretin | | Authors: | Palmieri, L.C, Freire, J.B.B, Palhano, F.L, Azevedo, E.P.C, Foguel, D, Lima, L.M.T.R. | | Deposit date: | 2009-07-10 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of human transthyretin variant A25T - #1
To be Published
|
|
2MWF
 
 | |
3W7A
 
 | | Crystal Structure of azoreductase AzrC fin complex with sulfone-modified azo dye Acid Red 88 | | Descriptor: | 4-[(E)-(2-hydroxynaphthalen-1-yl)diazenyl]naphthalene-1-sulfonic acid, CALCIUM ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yu, J, Ogata, D, Ooi, T, Yao, M. | | Deposit date: | 2013-02-27 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of AzrA and of AzrC complexed with substrate or inhibitor: insight into substrate specificity and catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
3IGK
 
 | | Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs (p53-DNA complex 2) | | Descriptor: | Cellular tumor antigen p53, DNA (5'-D(*CP*GP*GP*GP*CP*AP*TP*GP*CP*CP*CP*G)-3'), ZINC ION | | Authors: | Suad, O, Rabinovich, D, Rozenberg, H, Shakked, Z. | | Deposit date: | 2009-07-28 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs
Nat.Struct.Mol.Biol., 17, 2010
|
|
3CQR
 
 | |
6SYZ
 
 | | Crystal structure of YTHDC1 with fragment 1 (DHU_DC1_141) | | Descriptor: | SULFATE ION, YTH domain-containing protein 1, ~{N}-methylthieno[3,2-d]pyrimidin-4-amine | | Authors: | Bedi, R.K, Huang, D, Sledz, P, Caflisch, A. | | Deposit date: | 2019-10-01 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Selectively Disrupting m6A-Dependent Protein-RNA Interactions with Fragments.
Acs Chem.Biol., 15, 2020
|
|
