7XZF
 
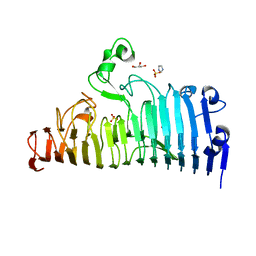 | | Wild type of the N-terminal domain of fucoidan lyase FdlA | | Descriptor: | Fucoidan lyase, GLYCEROL, IMIDAZOLE, ... | | Authors: | Wang, J, Li, M, Pan, X. | | Deposit date: | 2022-06-02 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural and Biochemical Analysis Reveals Catalytic Mechanism of Fucoidan Lyase from Flavobacterium sp. SA-0082.
Mar Drugs, 20, 2022
|
|
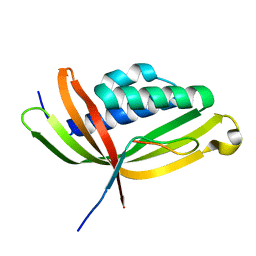
5GTB
 
 | |
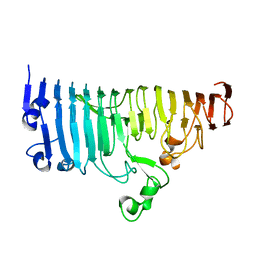
7XZ8
 
 | |
7XZB
 
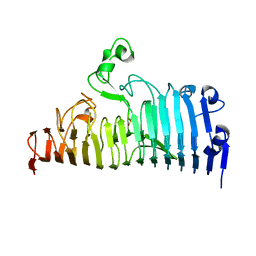 | |
7XZA
 
 | |
7XZ7
 
 | |
7XZE
 
 | |
7XZC
 
 | |
5HAD
 
 | |
4NSW
 
 | | Crystal structure of the BAR-PH domain of ACAP1 | | Descriptor: | Arf-GAP with coiled-coil, ANK repeat and PH domain-containing protein 1 | | Authors: | Pang, X, Zhang, K, Ma, J, Zhou, Q, Sun, F. | | Deposit date: | 2013-11-29 | | Release date: | 2014-10-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A PH Domain in ACAP1 Possesses Key Features of the BAR Domain in Promoting Membrane Curvature
Dev.Cell, 31, 2014
|
|
3HJ2
 
 | |
8TAG
 
 | | TMEM16F, with Calcium and PIP2, no inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Anoctamin-6, CALCIUM ION | | Authors: | Feng, S, Cheng, Y. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Identification of a drug binding pocket in TMEM16F calcium-activated ion channel and lipid scramblase.
Nat Commun, 14, 2023
|
|
4GB9
 
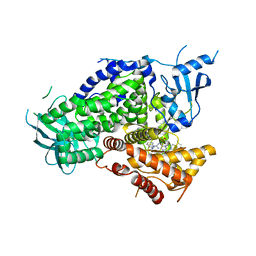 | | Potent and Highly Selective Benzimidazole Inhibitors of PI3K-delta | | Descriptor: | 2-[1-({2-[2-(dimethylamino)-1H-benzimidazol-1-yl]-9-methyl-6-(morpholin-4-yl)-9H-purin-8-yl}methyl)piperidin-4-yl]propan-2-ol, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M. | | Deposit date: | 2012-07-26 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.438 Å) | | Cite: | Potent and highly selective benzimidazole inhibitors of PI3-kinase delta.
J.Med.Chem., 55, 2012
|
|
4KY9
 
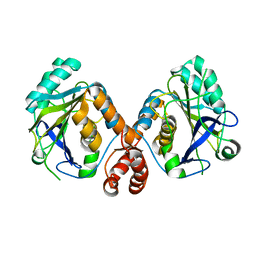 | |
7DNC
 
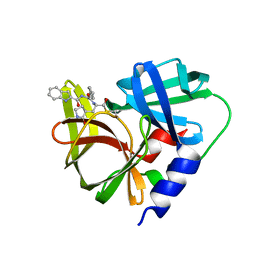 | | Crystal structure of EV71 3C proteinase in complex with a novel inhibitor | | Descriptor: | 3C protease, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Xie, H, Su, H.X, Li, M.J, Xu, Y.C. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Peptidomimetic Aldehydes as Broad-Spectrum Inhibitors against Enterovirus and SARS-CoV-2.
J.Med.Chem., 65, 2022
|
|
5EK0
 
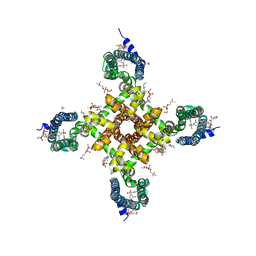 | | Human Nav1.7-VSD4-NavAb in complex with GX-936. | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-cyano-4-[2-[2-(1-ethylazetidin-3-yl)pyrazol-3-yl]-4-(trifluoromethyl)phenoxy]-~{N}-(1,2,4-thiadiazol-5-yl)benzenesulfonamide, Chimera of bacterial Ion transport protein and human Sodium channel protein type 9 subunit alpha | | Authors: | Ahuja, S, Mukund, S, Starovasnik, M.A, Koth, C.M, Payandeh, J. | | Deposit date: | 2015-11-03 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist.
Science, 350, 2015
|
|
6A8N
 
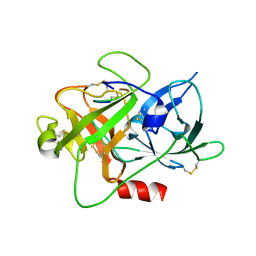 | | The crystal structure of muPAin-1-IG-2 in complex with muPA-SPD at pH8.5 | | Descriptor: | CYS-PRO-ALA-TYR-SER-ARG-TYR-ILE-GLY-CYS, Urokinase-type plasminogen activator B | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
6AKY
 
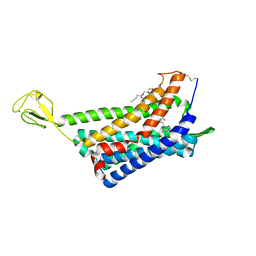 | | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 34 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4,4-difluoro-N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]octan-8-yl}-1-(thiophen-3-yl)propyl]cyclohexane-1-carboxamide, C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, ... | | Authors: | Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists.
J. Med. Chem., 61, 2018
|
|
7DL0
 
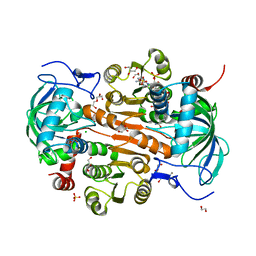 | | The mutant E310G/A314Y of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, ... | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL7
 
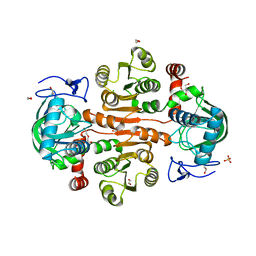 | | The wild-type structure of 3,5-DAHDHcca | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,5-diaminohexanoate dehydrogenase, ... | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-26 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.30065823 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL1
 
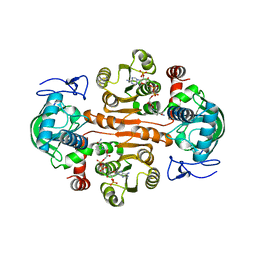 | | The mutant E310G/G323S structure of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 3,5-diaminohexanoate dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DL3
 
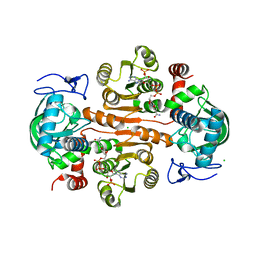 | | The structure of 3,5-DAHDHcca complex with NADPH | | Descriptor: | 3,5-diaminohexanoate dehydrogenase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Liu, N, Wu, L, Zhu, D.M, Zhou, J.H. | | Deposit date: | 2020-11-25 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84606934 Å) | | Cite: | Crystal Structures and Catalytic Mechanism of l-erythro-3,5-Diaminohexanoate Dehydrogenase and Rational Engineering for Asymmetric Synthesis of beta-Amino Acids.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5EWI
 
 | |
6AKX
 
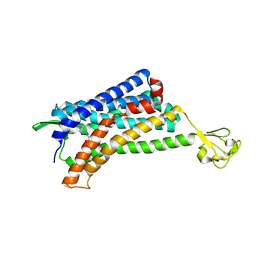 | | The Crystal structure of Human Chemokine Receptor CCR5 in complex with compound 21 | | Descriptor: | C-C chemokine receptor type 5,Rubredoxin,C-C chemokine receptor type 5, N-[(1S)-3-{(3-exo)-3-[3-methyl-5-(propan-2-yl)-4H-1,2,4-triazol-4-yl]-8-azabicyclo[3.2.1]octan-8-yl}-1-(thiophen-2-yl)propyl]cyclopentanecarboxamide, NITRATE ION, ... | | Authors: | Zhu, Y, Zhao, Q, Wu, B. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of 1-Heteroaryl-1,3-propanediamine Derivatives as a Novel Series of CC-Chemokine Receptor 5 Antagonists.
J. Med. Chem., 61, 2018
|
|
8T9D
 
 | | CryoEM structure of TR-TRAP | | Descriptor: | Mediator of RNA polymerase II transcription subunit 1, Mediator of RNA polymerase II transcription subunit 10, Mediator of RNA polymerase II transcription subunit 11, ... | | Authors: | Zhao, H, Asturias, F. | | Deposit date: | 2023-06-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.66 Å) | | Cite: | An IDR-dependent mechanism for nuclear receptor control of Mediator interaction with RNA polymerase II.
Mol.Cell, 84, 2024
|
|
