3SUV
 
 | |
3SUR
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with NAG-thiazoline. | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-hexosaminidase, SULFATE ION | | Authors: | Sumida, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-07-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Gaining insight into the inhibition of glycoside hydrolase family 20 exo-beta-N-acetylhexosaminidases using a structural approach
Org.Biomol.Chem., 10, 2012
|
|
3VQW
 
 | | Crystal structure of the SeMet substituted catalytic domain of pyrrolysyl-tRNA synthetase | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pyrrolysine--tRNA ligase | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-01 | | Release date: | 2013-01-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VQY
 
 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in complex with BocLys and AMPPNP (form 2) | | Descriptor: | MAGNESIUM ION, N~6~-(tert-butoxycarbonyl)-L-lysine, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-02 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3SUS
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with Gal-NAG-thiazoline | | Descriptor: | (3aR,5R,6R,7R,7aR)-5-(hydroxymethyl)-2-methyl-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, Beta-hexosaminidase, SULFATE ION | | Authors: | Sumida, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-07-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gaining insight into the inhibition of glycoside hydrolase family 20 exo-beta-N-acetylhexosaminidases using a structural approach
Org.Biomol.Chem., 10, 2012
|
|
3SUW
 
 | |
3SUT
 
 | |
3SUU
 
 | | Crystal structure of beta-hexosaminidase from Paenibacillus sp. TS12 in complex with Gal-PUGNAc | | Descriptor: | Beta-hexosaminidase, SULFATE ION, [(Z)-[(3R,4R,5R,6R)-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-ylidene]amino] N-phenylcarbamate | | Authors: | Sumida, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-07-11 | | Release date: | 2012-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gaining insight into the inhibition of glycoside hydrolase family 20 exo-beta-N-acetylhexosaminidases using a structural approach
Org.Biomol.Chem., 10, 2012
|
|
3VUZ
 
 | | Crystal structure of histone methyltransferase SET7/9 in complex with AAM-1 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl](hexyl)amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase SETD7 | | Authors: | Niwa, H, Handa, N, Tomabechi, Y, Honda, K, Toyama, M, Ohsawa, N, Shirouzu, M, Kagechika, H, Hirano, T, Umehara, T, Yokoyama, S. | | Deposit date: | 2012-07-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of histone methyltransferase SET7/9 in complexes with adenosylmethionine derivatives
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3VV0
 
 | | Crystal structure of histone methyltransferase SET7/9 in complex with DAAM-3 | | Descriptor: | 5'-{[(3S)-3-amino-3-carboxypropyl][2-(hexylamino)ethyl]amino}-5'-deoxyadenosine, Histone-lysine N-methyltransferase SETD7 | | Authors: | Niwa, H, Handa, N, Tomabechi, Y, Honda, K, Toyama, M, Ohsawa, N, Shirouzu, M, Kagechika, H, Hirano, T, Umehara, T, Yokoyama, S. | | Deposit date: | 2012-07-10 | | Release date: | 2013-03-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structures of histone methyltransferase SET7/9 in complexes with adenosylmethionine derivatives
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3UMW
 
 | | Crystal structure of Pim1 kinase in complex with inhibitor (Z)-2-[(1H-indazol-3-yl)methylene]-6-methoxy-7-(piperazin-1-ylmethyl)benzofuran-3(2H)-one | | Descriptor: | (2Z)-2-(1H-indazol-3-ylmethylidene)-6-methoxy-7-(piperazin-1-ylmethyl)-1-benzofuran-3(2H)-one, GLYCEROL, Proto-oncogene serine/threonine-protein kinase pim-1, ... | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2011-11-14 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Rational evolution of a novel type of potent and selective proviral integration site in Moloney murine leukemia virus kinase 1 (PIM1) inhibitor from a screening-hit compound.
J.Med.Chem., 55, 2012
|
|
3VYW
 
 | | Crystal structure of MNMC2 from Aquifex Aeolicus | | Descriptor: | BENZAMIDINE, MNMC2, S-ADENOSYLMETHIONINE | | Authors: | Shibata, R, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-10-03 | | Release date: | 2012-10-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization and structure of the Aquifex aeolicus protein DUF752: a bacterial tRNA-methyltransferase (MnmC2) functioning without the usually fused oxidase domain (MnmC1).
J.Biol.Chem., 287, 2012
|
|
3UMX
 
 | | Crystal structure of Pim1 kinase in complex with inhibitor (Z)-2-[(1H-indol-3-yl)methylene]-7-(azepan-1-ylmethyl)-6-hydroxybenzofuran-3(2H)-one | | Descriptor: | (2Z)-7-(azepan-1-ylmethyl)-6-hydroxy-2-(1H-indol-3-ylmethylidene)-1-benzofuran-3(2H)-one, Proto-oncogene serine/threonine-protein kinase pim-1, SULFATE ION | | Authors: | Parker, L.J, Handa, N, Yokoyama, S. | | Deposit date: | 2011-11-15 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Flexibility of the P-loop of Pim-1 kinase: observation of a novel conformation induced by interaction with an inhibitor
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3WU6
 
 | | Oxidized E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
3VTA
 
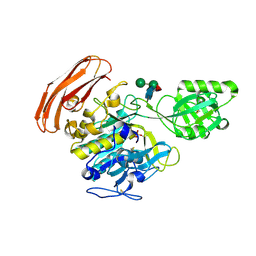 | | Crystal Structure of cucumisin, a subtilisin-like endoprotease from Cucumis melo L | | Descriptor: | Cucumisin, DIISOPROPYL PHOSPHONATE, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Murayama, K, Kato-Murayama, M, Hosaka, T, Sotokawauchi, A, Shirouzu, M, Arima, K, Yokoyama, S. | | Deposit date: | 2012-05-23 | | Release date: | 2012-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of cucumisin, a subtilisin-like endoprotease from Cucumis melo L
J.Mol.Biol., 423, 2012
|
|
3VHL
 
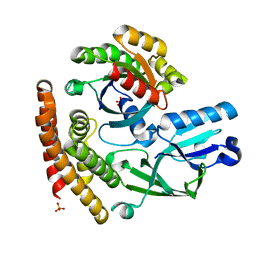 | | Crystal structure of the DHR-2 domain of DOCK8 in complex with Cdc42 (T17N mutant) | | Descriptor: | Cell division control protein 42 homolog, Dedicator of cytokinesis protein 8, PHOSPHATE ION | | Authors: | Hanawa-Suetsugu, K, Kukimoto-Niino, M, Nishizak, T, Terada, T, Shirouzu, M, Fukui, Y, Yokoyama, S. | | Deposit date: | 2011-08-26 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | DOCK8 is a Cdc42 activator critical for interstitial dendritic cell migration during immune responses.
Blood, 119, 2012
|
|
3VQX
 
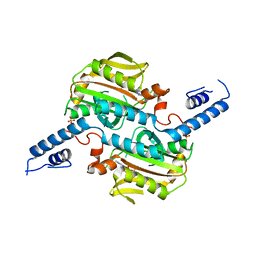 | | Crystal structure of the catalytic domain of pyrrolysyl-tRNA synthetase in triclinic crystal form | | Descriptor: | ADENOSINE MONOPHOSPHATE, PHOSPHATE ION, Pyrrolysine--tRNA ligase, ... | | Authors: | Yanagisawa, T, Sumida, T, Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2012-04-02 | | Release date: | 2013-01-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A novel crystal form of pyrrolysyl-tRNA synthetase reveals the pre- and post-aminoacyl-tRNA synthesis conformational states of the adenylate and aminoacyl moieties and an asparagine residue in the catalytic site
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3WPS
 
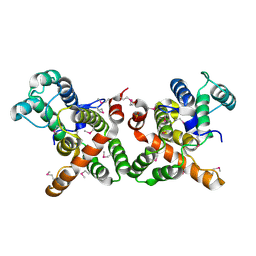 | | crystal structure of the GAP domain of MgcRacGAP(S387D) | | Descriptor: | Rac GTPase-activating protein 1, SULFATE ION | | Authors: | Murayama, K, Kato-murayama, M, Shirouzu, M, Kitamura, T, Yokoyama, S. | | Deposit date: | 2014-01-15 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | crystal structure of the GAP domain of MgcRacGAP
To be Published
|
|
3WPQ
 
 | |
3WHE
 
 | | A new conserved neutralizing epitope at the globular head of hemagglutinin in H3N2 influenza viruses | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fujii, Y, Sumida, T, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-08-25 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses.
J.Virol., 88, 2014
|
|
3WU5
 
 | | Reduced E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
3WJ9
 
 | |
3WU3
 
 | | Reduced-form structure of E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
3WU4
 
 | | Oxidized-form structure of E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
2DB3
 
 | | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa | | Descriptor: | 5'-R(*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ATP-dependent RNA helicase vasa, MAGNESIUM ION, ... | | Authors: | Sengoku, T, Nureki, O, Nakamura, A, Kobayashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-12-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa.
Cell(Cambridge,Mass.), 125, 2006
|
|
