8YL5
 
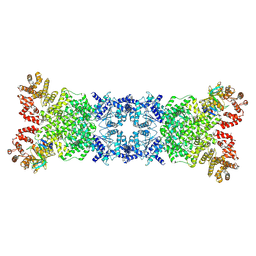 | | The DSR2-DSAD1 complex with DSAD1 on the same sides | | Descriptor: | DSAD1, SIR2-like domain-containing protein | | Authors: | Yang, X, Zheng, J. | | Deposit date: | 2024-03-05 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into autoinhibition and activation of defense-associated sirtuin protein.
Int.J.Biol.Macromol., 277, 2024
|
|
9C80
 
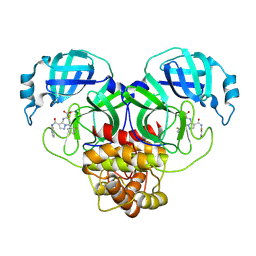 | | Co-structure of SARS-CoV-2 (COVID-19 with covalent inhibitor | | Descriptor: | (5R,7S,8R)-7-(2-fluorophenyl)-3-[(2-fluorophenyl)carbamoyl]-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-5-carboxylic acid, 3C-like proteinase nsp5 | | Authors: | Ornelas, E, Knapp, M.S. | | Deposit date: | 2024-06-11 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Identification of Potent, Broad-Spectrum Coronavirus Main Protease Inhibitors for Pandemic Preparedness.
J.Med.Chem., 67, 2024
|
|
9C8Q
 
 | |
8Z18
 
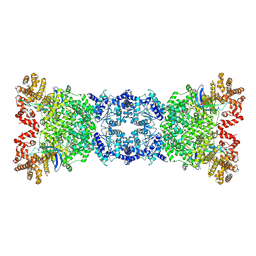 | |
9C7W
 
 | | human OC43 Main Protease (1-303) in complex with potent inhibitor | | Descriptor: | (8S)-3-(4,4-difluorocyclohexyl)-5-(pyrimidin-2-yl)pyrazolo[1,5-a]pyrimidine, ORF1ab polyprotein | | Authors: | Tang, J.Y, Knapp, M.S. | | Deposit date: | 2024-06-11 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Identification of Potent, Broad-Spectrum Coronavirus Main Protease Inhibitors for Pandemic Preparedness.
J.Med.Chem., 67, 2024
|
|
8YLT
 
 | | The structure of DSR2 and NAD+ complex | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SIR2-like domain-containing protein | | Authors: | Zheng, J, Yang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural insights into autoinhibition and activation of defense-associated sirtuin protein.
Int.J.Biol.Macromol., 277, 2024
|
|
9GMO
 
 | |
8YLN
 
 | | The structure of DSR2-Tail tube complex | | Descriptor: | Bacillus phage SPR Tube protein, SIR2-like domain-containing protein | | Authors: | Zheng, J, Yang, X. | | Deposit date: | 2024-03-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Structural insights into autoinhibition and activation of defense-associated sirtuin protein.
Int.J.Biol.Macromol., 277, 2024
|
|
8YKF
 
 | | The DSR2-DSAD1 complex with DSAD1 on the opposite sides | | Descriptor: | DSAD1, SIR2-like domain-containing protein | | Authors: | Zheng, J, Yang, X. | | Deposit date: | 2024-03-05 | | Release date: | 2024-08-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into autoinhibition and activation of defense-associated sirtuin protein.
Int.J.Biol.Macromol., 277, 2024
|
|
8ZTR
 
 | |
7YS6
 
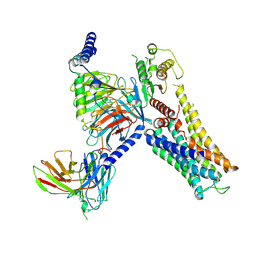 | | Cryo-EM structure of the Serotonin 6 (5-HT6) receptor-DNGs-scFv16 complex | | Descriptor: | 5-hydroxytryptamine receptor 6, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, Q.Y, Wang, Y.F, He, L, Wang, S, Cong, Y. | | Deposit date: | 2022-08-11 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into constitutive activity of 5-HT 6 receptor.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7YUJ
 
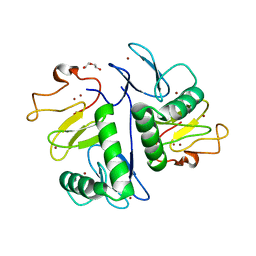 | | Crystal structure of HOIL-1L(365-510) | | Descriptor: | DI(HYDROXYETHYL)ETHER, RanBP-type and C3HC4-type zinc finger-containing protein 1, ZINC ION | | Authors: | Xiao, L, Pan, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Mechanistic insights into the enzymatic activity of E3 ligase HOIL-1L and its regulation by the linear ubiquitin chain binding.
Sci Adv, 9, 2023
|
|
7YUI
 
 | |
6OSY
 
 | |
6LJ9
 
 | |
6XP5
 
 | | Head-Middle module of Mediator | | Descriptor: | HEAT, Med22, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Zhang, H.Q, Chen, D.C, Kornberg, R.D. | | Deposit date: | 2020-07-08 | | Release date: | 2021-03-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mediator structure and conformation change.
Mol.Cell, 81, 2021
|
|
4WWI
 
 | |
6LJB
 
 | |
6PM9
 
 | | Crystal structure of the core catalytic domain of human O-GlcNAcase bound to MK-8719 | | Descriptor: | (3aR,5S,6S,7R,7aR)-5-(difluoromethyl)-2-(ethylamino)-5,6,7,7a-tetrahydro-3aH-pyrano[3,2-d][1,3]thiazole-6,7-diol, O-GlcNAcase TIM-barrel domain, O-GlcNAcase stalk domain | | Authors: | Klein, D.J, Selnick, H.G, Duffy, J.L, McEachern, E.J. | | Deposit date: | 2019-07-01 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of MK-8719, a Potent O-GlcNAcase Inhibitor as a Potential Treatment for Tauopathies.
J.Med.Chem., 62, 2019
|
|
4WXX
 
 | | The crystal structure of human DNMT1(351-1600) | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Zhang, Z.M, Song, J. | | Deposit date: | 2014-11-14 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.622 Å) | | Cite: | Crystal Structure of Human DNA Methyltransferase 1.
J.Mol.Biol., 427, 2015
|
|
7PG9
 
 | | human 20S proteasome | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-08-13 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The 20S as a stand-alone proteasome in cells can degrade the ubiquitin tag.
Nat Commun, 12, 2021
|
|
6OT1
 
 | |
6KJW
 
 | | Galectin-13 variant C136S | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Galectin-13/placental protein 13: redox-active disulfides as switches for regulating structure, function and cellular distribution.
Glycobiology, 30, 2020
|
|
5JDR
 
 | | Structure of PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Zhou, A, Wei, H. | | Deposit date: | 2016-04-17 | | Release date: | 2017-04-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of a novel PD-L1 nanobody for immune checkpoint blockade.
Cell Discov, 3, 2017
|
|
4MBI
 
 | | Discovery of Pyrazolo[1,5a]pyrimidine-based Pim1 Inhibitors | | Descriptor: | N,N-dimethyl-N'-[3-(1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidin-5-yl]ethane-1,2-diamine, Serine/threonine-protein kinase pim-1 | | Authors: | Azevedo, R, Fischmann, T.O. | | Deposit date: | 2013-08-19 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of pyrazolo[1,5-a]pyrimidine-based Pim inhibitors: A template-based approach.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
