7WCZ
 
 | | SARS-CoV-2 Beta spike in complex with one S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCR
 
 | | RBD-1 of SARS-CoV-2 Beta spike in complex with S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WEV
 
 | | SARS-COV-2 BETA VARIANT SPIKE PROTEIN IN TRANSITION STATE | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2021-12-24 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Conformational dynamics of the Beta and Kappa SARS-CoV-2 spike proteins and their complexes with ACE2 receptor revealed by cryo-EM.
Nat Commun, 12, 2021
|
|
7WR4
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex | | Descriptor: | Calmodulin-1, Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR2
 
 | |
7WR0
 
 | | P32 of caspase-4 C258A mutant | | Descriptor: | Caspase-4 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR6
 
 | | Crystal structure of ADP-riboxanated caspase-4 in complex with Af1521 | | Descriptor: | ADP-ribose glycohydrolase AF_1521, Caspase-4, [[(3~{a}~{S},5~{R},6~{R},6~{a}~{R})-2-azanylidene-3-[(4~{R})-4-azanyl-5-oxidanylidene-pentyl]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxazol-5-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR1
 
 | | P32 of caspase-4 C258A mutant in complex with OspC3 C-terminal ankyrin-repeat domain | | Descriptor: | Caspase-4, OspC3 | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR3
 
 | | Crystal structure of MBP-fused OspC3 in complex with calmodulin | | Descriptor: | Calmodulin-1, MBP-fused OspC3, NICOTINAMIDE, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WR5
 
 | | Crystal structure of OspC3-calmodulin-caspase-4 complex binding with 2'-aF-NAD+ | | Descriptor: | Calmodulin-1, Caspase-4, OspC3, ... | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7WZ5
 
 | | Larimichthys crocea IFNi | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon C | | Authors: | Chen, J.J. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-08 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural and Functional Characterization of a Fish Type I Subgroup d IFN Reveals Its Binding to Receptors.
J Immunol., 212, 2024
|
|
7X3L
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07044 | | Descriptor: | (2~{R})-2-[4-(3-fluoranyl-4-methyl-phenyl)-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7X3M
 
 | | Crystal structure of Aldo-keto reductase 1C3 complexed with compound S07045 | | Descriptor: | (2~{R})-2-[4-[3,5-bis(chloranyl)phenyl]-3-(trifluoromethyl)phenyl]butanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Jiang, J, Liu, Y, He, S, Chen, Y, Chu, X, Liu, Y, Guo, Q, Zhao, L, Feng, F, Liu, W, Zhang, X, Fang, P, Sun, H. | | Deposit date: | 2022-03-01 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.694 Å) | | Cite: | Development of Biaryl-Containing Aldo-Keto Reductase 1C3 (AKR1C3) Inhibitors for Reversing AKR1C3-Mediated Drug Resistance in Cancer Treatment.
J.Med.Chem., 66, 2023
|
|
7WZS
 
 | |
7FEF
 
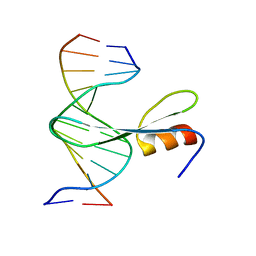 | | Crystal structure of AtMBD6 with DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain-containing protein 6 | | Authors: | Wu, Z.B, Liu, K, Min, J.R. | | Deposit date: | 2021-07-19 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
7FEO
 
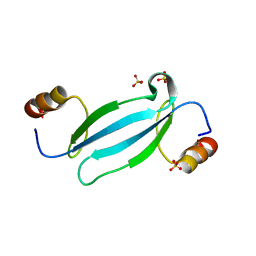 | | Crystal structure of AtMBD5 MBD domain | | Descriptor: | Methyl-CpG-binding domain-containing protein 5, SULFATE ION | | Authors: | Zhou, M.Q, Wu, Z.B, Liu, K, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-21 | | Release date: | 2021-12-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Family-wide Characterization of Methylated DNA Binding Ability of Arabidopsis MBDs.
J.Mol.Biol., 434, 2022
|
|
8Y9N
 
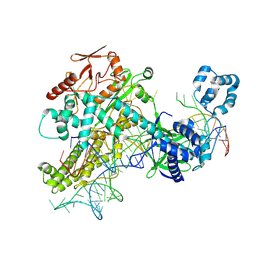 | | Cas12h1-crRNA-dsDNA ternary complex | | Descriptor: | Cas12h1, DNA (29-mer), MAGNESIUM ION, ... | | Authors: | Zheng, W.W, Liu, M.X. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular insights and rational engineering of a compact CRISPR-Cas effector Cas12h1 with a broad-spectrum PAM.
Signal Transduct Target Ther, 10, 2025
|
|
8Y9M
 
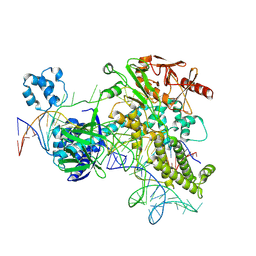 | | Cas12h1(D465A)-crRNA-dsDNA ternary complex | | Descriptor: | Cas12h1 (D465A) mutant, DNA (29-mer), crRNA | | Authors: | Zheng, W.W, Liu, M.X. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Molecular insights and rational engineering of a compact CRISPR-Cas effector Cas12h1 with a broad-spectrum PAM.
Signal Transduct Target Ther, 10, 2025
|
|
8Y9L
 
 | | Cas12h1-crRNA binary complex | | Descriptor: | Cas12h1, MAGNESIUM ION, crRNA | | Authors: | Zheng, W.W, Liu, M.X. | | Deposit date: | 2024-02-07 | | Release date: | 2025-02-12 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular insights and rational engineering of a compact CRISPR-Cas effector Cas12h1 with a broad-spectrum PAM.
Signal Transduct Target Ther, 10, 2025
|
|
8YHX
 
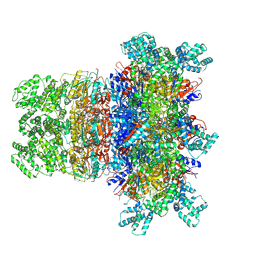 | | Cryo-EM structure of the trimeric HerA | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, DUF87 domain-containing protein, SIR2 family protein | | Authors: | Zhen, X, Zhou, B, Xiong, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Mechanistic basis for the allosteric activation of NADase activity in the Sir2-HerA antiphage defense system.
Nat Commun, 15, 2024
|
|
8YHO
 
 | | Cryo-EM structure of the trimeric HerA | | Descriptor: | DUF87 domain-containing protein | | Authors: | Zhen, X, Xiong, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanistic basis for the allosteric activation of NADase activity in the Sir2-HerA antiphage defense system.
Nat Commun, 15, 2024
|
|
9AV4
 
 | |
9AV5
 
 | |
9AV8
 
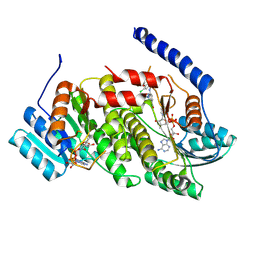 | |
8JPX
 
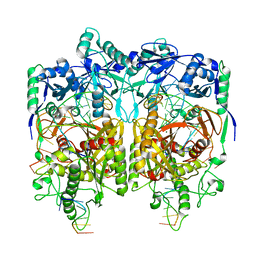 | | Cryo-EM structure of PfAgo-guide DNA-target DNA complex | | Descriptor: | Excess DNA, Guide DNA, MAGNESIUM ION, ... | | Authors: | Zhuang, L. | | Deposit date: | 2023-06-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular mechanism for target recognition, dimerization, and activation of Pyrococcus furiosus Argonaute.
Mol.Cell, 84, 2024
|
|
