5XAD
 
 | | NLIR - LC3B fusion protein | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, Uncharacterised protein | | Authors: | Kwon, D.H, Kim, L, Song, H.K. | | Deposit date: | 2017-03-12 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A novel conformation of the LC3-interacting region motif revealed by the structure of a complex between LC3B and RavZ
Biochem. Biophys. Res. Commun., 490, 2017
|
|
4B22
 
 | | Unprecedented sculpting of DNA at abasic sites by DNA glycosylase homolog Mag2 | | Descriptor: | 5'-D(*CP*GP*AP*TP*GP*GP*GP*TP*AP*GP*CP)-3', 5'-D(*GP*CP*TP*AP*CP*(3DR)P*CP*AP*TP*CP*GP)-3', MAG2, ... | | Authors: | Dalhus, B, Nilsen, L, Korvald, H, Huffman, J, Forstrom, R.J, McMurray, C.T, Alseth, I, Tainer, J.A, Bjoras, M. | | Deposit date: | 2012-07-12 | | Release date: | 2013-01-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Sculpting of DNA at Abasic Sites by DNA Glycosylase Homolog Mag2.
Structure, 21, 2013
|
|
6GBQ
 
 | | Crystal Structure of the oligomerization domain of Vp35 from Reston virus | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Zinzula, L, Nagy, I, Orsini, M, Weyher-Stingl, E, Baumeister, W, Bracher, A. | | Deposit date: | 2018-04-16 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structures of Ebola and Reston Virus VP35 Oligomerization Domains and Comparative Biophysical Characterization in All Ebolavirus Species.
Structure, 27, 2019
|
|
5XBM
 
 | | Structure of SCARB2-JL2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhang, X, Yang, P, Wang, N, Zhang, J, Li, J, Guo, H, Yin, X, Rao, Z, Wang, X, Zhang, L. | | Deposit date: | 2017-03-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | The binding of a monoclonal antibody to the apical region of SCARB2 blocks EV71 infection.
Protein Cell, 8, 2017
|
|
3TZZ
 
 | | Crystal structure of a fragment containing the acyltransferase domain of Pks13 from Mycobacterium tuberculosis in the carboxypalmitoylated form at 2.5 A | | Descriptor: | 12-mer peptide, 2-(2-METHOXYETHOXY)ETHANOL, GLYCEROL, ... | | Authors: | Bergeret, F, Pedelacq, J.D, Mourey, L, Bon, C. | | Deposit date: | 2011-09-28 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Biochemical and structural study of the atypical acyltransferase domain from the mycobacterial polyketide synthase pks13
J.Biol.Chem., 287, 2012
|
|
1DOL
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, I-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1 | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A. | | Deposit date: | 1996-11-22 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
6G0H
 
 | |
8X4F
 
 | |
1DQ1
 
 | | Calcium;Calcium concanavalin A | | Descriptor: | CALCIUM ION, Concanavalin-Br | | Authors: | Bouckaert, J, Dewallef, Y, Poortmans, F, Wyns, L, Loris, R. | | Deposit date: | 1999-12-29 | | Release date: | 2000-01-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural features of concanavalin A governing non-proline peptide isomerization
J.Biol.Chem., 275, 2000
|
|
8X7X
 
 | | Crystal structure of SADS-CoV fusion core | | Descriptor: | CHLORIDE ION, HR1, HR2 | | Authors: | Yan, L. | | Deposit date: | 2023-11-26 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Fusion Cores from CCoV-HuPn-2018 and SADS-CoV.
Viruses, 16, 2024
|
|
3U1M
 
 | | Structure of the mRNA splicing complex component Cwc2 | | Descriptor: | Pre-mRNA-splicing factor CWC2, ZINC ION | | Authors: | Lu, P, Lu, G, Yan, C, Wang, L, Li, W, Yin, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the mRNA splicing complex component Cwc2: insights into RNA recognition
Biochem.J., 441, 2012
|
|
1DSW
 
 | | THE SOLUTION STRUCTURE OF A MONOMERIC, REDUCED FORM OF HUMAN COPPER, ZINC SUPEROXIDE DISMUTASE BEARING THE SAME CHARGE AS THE NATIVE PROTEIN | | Descriptor: | COPPER (II) ION, SUPEROXIDE DISMUTASE (CU-ZN), ZINC ION | | Authors: | Banci, L, Bertini, I, Del Conte, R, Fadin, R, Mangani, S, Viezzoli, M.S. | | Deposit date: | 2000-01-10 | | Release date: | 2000-03-22 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | The solution structure of a monomeric, reduced form of human copper,zinc superoxide dismutase bearing the same charge as the native protein.
J.Biol.Inorg.Chem., 4, 1999
|
|
3U2H
 
 | | Crystal structure of the C-terminal DUF1608 domain of the Methanosarcina acetivorans S-layer (MA0829) protein | | Descriptor: | GLYCEROL, S-layer protein MA0829 | | Authors: | Chan, S, Phan, T, Ahn, C.J, Shin, A, Rohlin, L, Gunsalus, R.P, Arbing, M.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structure of the surface layer of the methanogenic archaean Methanosarcina acetivorans.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8T7C
 
 | | Crystal structure of human phospholipase C gamma 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2, CALCIUM ION | | Authors: | Chen, Y, Choi, H, Zhuang, N, Hu, L, Qian, D, Wang, J. | | Deposit date: | 2023-06-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal and cryo-EM structures of PLCg2 reveal dynamic inter-domain recognitions in autoinhibition
To Be Published
|
|
6G48
 
 | | Sporosarcina pasteurii urease inhibited by silver | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, NICKEL (II) ION, ... | | Authors: | Mazzei, L, Cianci, M, Ciurli, S. | | Deposit date: | 2018-03-27 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The structure of urease inactivated by Ag(i): a new paradigm for enzyme inhibition by heavy metals.
Dalton Trans, 47, 2018
|
|
1PDV
 
 | |
4B82
 
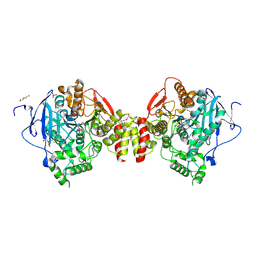 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino- ethyl)-2-fluoranyl-benzenesulfonamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4B84
 
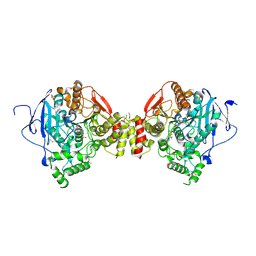 | | Mus musculus Acetylcholinesterase in complex with N-(2-Diethylamino- ethyl)-3-trifluoromethyl-benzenesulfonamide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15-PENTAOXAHEPTADECANE, ACETYLCHOLINESTERASE, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
4BJ9
 
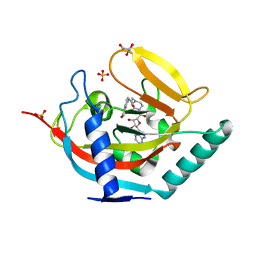 | | Crystal structure of human tankyrase 2 in complex with EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2013-04-17 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Evaluation and Structural Basis for the Inhibition of Tankyrases by Parp Inhibitors
Acs Med.Chem.Lett., 5, 2014
|
|
6G7M
 
 | | Four-site variant (Y222C, C197S, C432S, C433S) of E. coli hydrogenase-2 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, Hydrogenase-2 large chain, ... | | Authors: | Carr, S.B, Armstrong, F.A, Zhang, L, Beaton, S.E. | | Deposit date: | 2018-04-06 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Direct visible light activation of a surface cysteine-engineered [NiFe]-hydrogenase by silver nanoclusters
Energy Environ Sci, 2019
|
|
8CZV
 
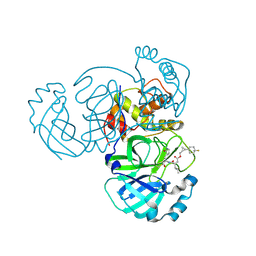 | | Structure of MERS 3CL protease in complex with the cyclopropane based inhibitor 17d | | Descriptor: | 3C-like proteinase, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, [(1~{S},2~{R})-2-[4,4-bis(fluoranyl)cyclohexyl]cyclopropyl]methyl ~{N}-[(2~{S})-1-[[(1~{S},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Liu, L, Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
4B81
 
 | | Mus musculus Acetylcholinesterase in complex with 1-(4-Chloro-phenyl)- N-(2-diethylamino-ethyl)-methanesulfonamide | | Descriptor: | 1-(4-chlorophenyl)-N-[2-(diethylamino)ethyl]methanesulfonamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Andersson, C.D, Forsgren, N, Akfur, C, Allgardsson, A, Berg, L, Qian, W, Ekstrom, F, Linusson, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-09-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Divergent Structure-Activity Relationships of Structurally Similar Acetylcholinesterase Inhibitors.
J.Med.Chem., 56, 2013
|
|
3UBH
 
 | | Crystal structure of Drosophila N-cadherin EC1-4 | | Descriptor: | CALCIUM ION, Neural-cadherin | | Authors: | Jin, X, Walker, M.A, Shapiro, L. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Drosophila N-cadherin ectodomain regions reveal a widely used class of Ca2+-free interdomain linkers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5XKC
 
 | | Crystal structure of dibenzothiophene sulfone monooxygenase BdsA at 2.2 angstrome | | Descriptor: | Dibenzothiophene desulfurization enzyme A | | Authors: | Gu, L, Su, T, Liu, S, Su, J. | | Deposit date: | 2017-05-07 | | Release date: | 2018-05-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Structural and Biochemical Characterization of BdsA fromBacillus subtilisWU-S2B, a Key Enzyme in the "4S" Desulfurization Pathway.
Front Microbiol, 9, 2018
|
|
4BE7
 
 | | MUTANT (K220R) OF THE HSDR SUBUNIT OF THE ECOR124I RESTRICTION ENZYME IN COMPLEX WITH ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Csefalvay, E, Lapkouski, M, Guzanova, A, Csefalvay, L, Baikova, T, Shevelev, I, Janscak, P, Smatanova, I.K, Panjikar, S, Carey, J, Weiserova, M, Ettrich, R. | | Deposit date: | 2013-03-06 | | Release date: | 2014-03-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.744 Å) | | Cite: | Functional Coupling of Duplex Translocation to DNA Cleavage in a Type I Restriction Enzyme.
Plos One, 10, 2015
|
|
