2EOP
 
 | | Solution structure of the C2H2 type zinc finger (region 719-751) of human Zinc finger protein 268 | | Descriptor: | ZINC ION, Zinc finger protein 268 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 719-751) of human Zinc finger protein 268
To be Published
|
|
2EP3
 
 | | Solution structure of the C2H2 type zinc finger (region 631-663) of human Zinc finger protein 484 | | Descriptor: | ZINC ION, Zinc finger protein 484 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 631-663) of human Zinc finger protein 484
To be Published
|
|
5UFH
 
 | | The crystal structure of a LacI-type transcription regulator from Bifidobacterium animalis subsp. lactis DSM 10140 | | Descriptor: | GLYCEROL, LacI-type transcriptional regulator, NITRATE ION | | Authors: | Tan, K, Li, H, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-01-04 | | Release date: | 2017-01-18 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of a LacI-type transcription regulator from Bifidobacterium animalis subsp. lactis DSM 10140
To Be Published
|
|
5UHJ
 
 | | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234 | | Descriptor: | FORMIC ACID, Glyoxalase/bleomycin resisance protein/dioxygenase | | Authors: | Tan, K, Li, H, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-11 | | Release date: | 2017-01-25 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of a natural product biosynthetic enzyme from Streptomyces sp. CB03234
To Be Published
|
|
2EOH
 
 | | Solution structure of the C2H2 type zinc finger (region 780-812) of human Zinc finger protein 28 homolog | | Descriptor: | ZINC ION, Zinc finger protein 28 homolog | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 780-812) of human Zinc finger protein 28 homolog
To be Published
|
|
2EOU
 
 | | Solution structure of the C2H2 type zinc finger (region 370-400) of human Zinc finger protein 473 | | Descriptor: | ZINC ION, Zinc finger protein 473 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 370-400) of human Zinc finger protein 473
To be Published
|
|
2EOK
 
 | | Solution structure of the C2H2 type zinc finger (region 441-469) of human Zinc finger protein 268 | | Descriptor: | ZINC ION, Zinc finger protein 268 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 441-469) of human Zinc finger protein 268
To be Published
|
|
2EOV
 
 | | Solution structure of the C2H2 type zinc finger (region 519-551) of human Zinc finger protein 484 | | Descriptor: | ZINC ION, Zinc finger protein 484 | | Authors: | Tochio, N, Tomizawa, T, Abe, H, Saito, K, Li, H, Sato, M, Koshiba, S, Kobayashi, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C2H2 type zinc finger (region 519-551) of human Zinc finger protein 484
To be Published
|
|
5UJM
 
 | | Structure of the active form of human Origin Recognition Complex and its ATPase motor module | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Origin recognition complex subunit 1, ... | | Authors: | Tocilj, A, On, K, Yuan, Z, Sun, J, Elkayam, E, Li, H, Stillman, B, Joshua-Tor, L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
3Q6G
 
 | | Crystal structure of Fab of rhesus mAb 2.5B specific for quaternary neutralizing epitope of HIV-1 gp120 | | Descriptor: | Heavy chain of Fab of rhesus mAb 2.5B, Light chain of Fab of rhesus mAb 2.5B | | Authors: | Spurrier, B, Sampson, J, Totrov, M, Li, H, O'Neal, T, William, C, Robinson, J, Gorny, M.K, Zolla-Pazner, S, Kong, X.P. | | Deposit date: | 2010-12-31 | | Release date: | 2011-05-25 | | Last modified: | 2013-03-20 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structural Analysis of Human and Macaque mAbs 2909 and 2.5B: Implications for the Configuration of the Quaternary Neutralizing Epitope of HIV-1 gp120.
Structure, 19, 2011
|
|
3QOK
 
 | |
1OZE
 
 | | Crystal Structures of the Ferric, Ferrous, and Ferrous-NO Forms of the Asp140Ala Mutant of Human Heme Oxygenase-1:Catalytic Implications | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lad, L, Wang, J, Li, H, Friedman, J, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-04-08 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structures of the ferric, ferrous, and ferrous-NO forms of the Asp140Ala mutant of human heme oxygenase-1: catalytic implications
J.Mol.Biol., 330, 2003
|
|
3PKM
 
 | | Crystal structure of Cas6 with its substrate RNA | | Descriptor: | 5'-R(*AP*UP*UP*AP*CP*AP*AP*UP*AP*A)-3', 5'-R(P*UP*UP*AP*CP*AP*AP*UP*AP*A)-3', CRISPR-associated endoribonuclease Cas6 | | Authors: | Wang, R, Preamplume, G, Li, H. | | Deposit date: | 2010-11-11 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.103 Å) | | Cite: | Interaction of the Cas6 Riboendonuclease with CRISPR RNAs: Recognition and Cleavage.
Structure, 19, 2011
|
|
1OYL
 
 | | Crystal Structures of the Ferric, Ferrous, and Ferrous-NO Forms of the Asp140Ala Mutant of Human Heme Oxygenase-1: Catalytic Implications | | Descriptor: | Heme oxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Lad, L, Wang, J, Li, H, Friedman, J, Ortiz de Montellano, P.R, Poulos, T.L. | | Deposit date: | 2003-04-05 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Crystal structures of the ferric, ferrous, and ferrous-NO forms of the Asp140Ala mutant of human heme oxygenase-1: catalytic implications
J.Mol.Biol., 330, 2003
|
|
1OQR
 
 | | Crystal structure of C73S mutant of putidaredoxin, a [2Fe-2S] ferredoxin from Pseudomonas putida, at 1.65A resolution | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Sevrioukova, I.F, Garcia, C, Li, H, Bhaskar, B, Poulos, T.L. | | Deposit date: | 2003-03-10 | | Release date: | 2003-11-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of putidaredoxin, the [2Fe-2S] component of the P450cam monooxygenase system from Pseudomonas putida
J.MOL.BIOL., 333, 2003
|
|
1PKK
 
 | | Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Bifunctional deaminase/diphosphatase | | Authors: | Huffman, J.L, Li, H, White, R.H, Tainer, J.A. | | Deposit date: | 2003-06-05 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii
J.Mol.Biol., 331, 2003
|
|
3OOS
 
 | | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne | | Descriptor: | Alpha/beta hydrolase family protein, GLYCEROL, SULFATE ION, ... | | Authors: | Fan, Y, Tan, K, Bigelow, L, Hamilton, J, Li, H, Zhou, Y, Clancy, S, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-11-10 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The structure of an alpha/beta fold family hydrolase from Bacillus anthracis str. Sterne
To be Published
|
|
5URA
 
 | | Enantiomer-Specific Binding of the Potent Antinociceptive Agent SBFI-26 to Anandamide transporters FABP7 | | Descriptor: | (1S,2S,3S,4S)-3-{[(naphthalen-1-yl)oxy]carbonyl}-2,4-diphenylcyclobutane-1-carboxylic acid, Fatty acid-binding protein, brain, ... | | Authors: | Hsu, H.-C, Li, H. | | Deposit date: | 2017-02-09 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85002172 Å) | | Cite: | The Antinociceptive Agent SBFI-26 Binds to Anandamide Transporters FABP5 and FABP7 at Two Different Sites.
Biochemistry, 56, 2017
|
|
5UOA
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 3-[(2-Amino-4-methylquinolin-7-yl)methoxy]-5-((methylamino)methyl)benzonitrile | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(2-amino-4-methylquinolin-7-yl)methoxy]-5-[(methylamino)methyl]benzonitrile, CHLORIDE ION, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2017-01-31 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nitrile in the Hole: Discovery of a Small Auxiliary Pocket in Neuronal Nitric Oxide Synthase Leading to the Development of Potent and Selective 2-Aminoquinoline Inhibitors.
J. Med. Chem., 60, 2017
|
|
5UOU
 
 | | High resolution structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578 | | Descriptor: | 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) decarboxylase | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-01 | | Release date: | 2017-02-15 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High resolution structure of 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase from Klebsiella pneumoniae subsp. pneumoniae MGH 78578
To Be Published
|
|
4KCS
 
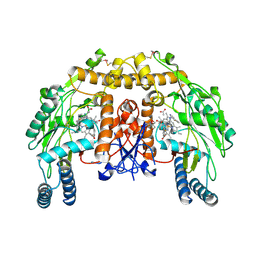 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with N-(3-((ethyl(3-fluorophenethyl)amino)methyl)phenyl)thiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, GLYCEROL, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent and Selective Double-Headed Thiophene-2-carboximidamide Inhibitors of Neuronal Nitric Oxide Synthase for the Treatment of Melanoma.
J.Med.Chem., 57, 2014
|
|
3OI8
 
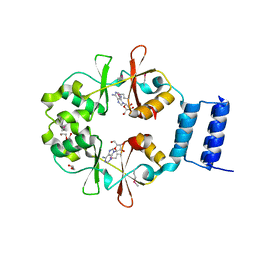 | | The crystal structure of functionally unknown conserved protein domain from Neisseria meningitidis MC58 | | Descriptor: | ADENOSINE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Zhang, R, Tan, K, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-18 | | Release date: | 2010-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | The crystal structure of functionally unknown conserved protein domain from Neisseria meningitidis MC58
To be Published
|
|
3OP1
 
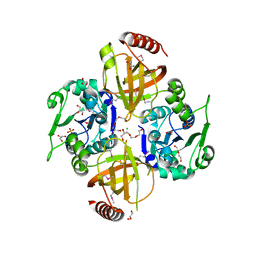 | | Crystal Structure of Macrolide-efflux Protein SP_1110 from Streptococcus pneumoniae | | Descriptor: | ACETIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Li, H, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.488 Å) | | Cite: | Crystal Structure of Macrolide-efflux Protein SP_1110 from Streptococcus pneumoniae
TO BE PUBLISHED
|
|
5V8F
 
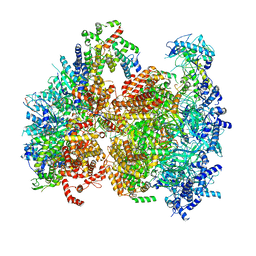 | | Structural basis of MCM2-7 replicative helicase loading by ORC-Cdc6 and Cdt1 | | Descriptor: | Cell division control protein 6, Cell division cycle protein CDT1, DNA (39-MER), ... | | Authors: | Yuan, Z, Riera, A, Bai, L, Sun, J, Spanos, C, Chen, Z.A, Barbon, M, Rappsilber, J, Stillman, B, Speck, C, Li, H. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-10 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1Q1R
 
 | |
