3IPE
 
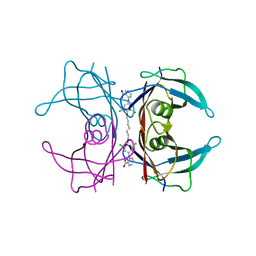 | | Human Transthyretin (TTR) complexed with a palindromic bivalent amyloid inhibitor (7 carbon linker). | | Descriptor: | 2,2'-{heptane-1,7-diylbis[oxy(3,5-dichlorobenzene-4,1-diyl)imino]}dibenzoic acid, Transthyretin | | Authors: | Kolstoe, S.E, Wood, S.P, Pepys, M.B. | | Deposit date: | 2009-08-17 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Trapping of palindromic ligands within native transthyretin prevents amyloid formation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4N6H
 
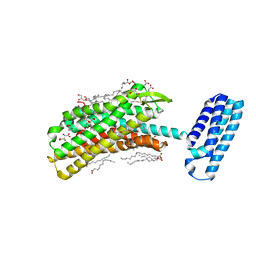 | | 1.8 A Structure of the human delta opioid 7TM receptor (PSI Community Target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4bS,8R,8aS,14bR)-7-(cyclopropylmethyl)-5,6,7,8,14,14b-hexahydro-4,8-methano[1]benzofuro[2,3-a]pyrido[4,3-b]carbazole-1,8a(9H)-diol, L(+)-TARTARIC ACID, ... | | Authors: | Fenalti, G, Giguere, P.M, Katritch, V, Huang, X.-P, Thompson, A.A, Han, G.W, Cherezov, V, Roth, B.L, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2013-10-12 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular control of delta-opioid receptor signalling.
Nature, 506, 2014
|
|
3HSA
 
 | |
3IRB
 
 | |
3I4B
 
 | | Crystal structure of GSK3b in complex with a pyrimidylpyrrole inhibitor | | Descriptor: | Glycogen synthase kinase-3 beta, N-[(1S)-2-hydroxy-1-phenylethyl]-4-[5-methyl-2-(phenylamino)pyrimidin-4-yl]-1H-pyrrole-2-carboxamide | | Authors: | Ter Haar, E. | | Deposit date: | 2009-07-01 | | Release date: | 2010-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control.
J.Med.Chem., 52, 2009
|
|
3HWK
 
 | |
3ICO
 
 | |
3H0N
 
 | |
3GVC
 
 | |
3I60
 
 | |
3GWA
 
 | |
3H7F
 
 | |
3HBZ
 
 | |
3GWE
 
 | |
3HE2
 
 | |
3JZF
 
 | |
3K5J
 
 | |
3HZG
 
 | | Crystal structure of mycobacterium tuberculosis thymidylate synthase X bound with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Staker, B.L, Rathod, P, Hunter, J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Increasing the structural coverage of tuberculosis drug targets.
Tuberculosis (Edinb), 95, 2015
|
|
3I5Z
 
 | |
3HWI
 
 | |
1E67
 
 | | Zn-Azurin from Pseudomonas aeruginosa | | Descriptor: | AZURIN, NITRATE ION, ZINC ION | | Authors: | Nar, H, Messerschmidt, A. | | Deposit date: | 2000-08-09 | | Release date: | 2000-08-16 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Characterization and Crystal Structure of Zinc Azurin, a by-Product of Heterologous Expression in Escherichia Coli of Pseudomonas Aeruginosa Copper Azurin
Eur.J.Biochem., 205, 1992
|
|
1E5Y
 
 | |
1D7E
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 1999-10-17 | | Release date: | 2000-03-31 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Conformational substates in different crystal forms of the photoactive yellow protein--correlation with theoretical and experimental flexibility.
Protein Sci., 9, 2000
|
|
3MBM
 
 | | Crystal structure of 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase from Burkholderia pseudomallei with cytosine and FoL fragment 717, imidazo[2,1-b][1,3]thiazol-6-ylmethanol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, 6-AMINOPYRIMIDIN-2(1H)-ONE, ... | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2010-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Leveraging structure determination with fragment screening for infectious disease drug targets: MECP synthase from Burkholderia pseudomallei.
J Struct Funct Genomics, 12, 2011
|
|
1E5Z
 
 | |
