9ODS
 
 | | Structure of CRBN TBD bound to compound C3 | | Descriptor: | (1P)-1-[(4S)-8H-spiro[furo[2,3-c]imidazo[1,2-a]pyridine-7,4'-piperidin]-3-yl]pyrimidine-2,4(1H,3H)-dione, Protein cereblon, ZINC ION | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2025-04-27 | | Release date: | 2025-05-28 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery and Characterization of PVTX-321 as a Potent and Orally Bioavailable Estrogen Receptor Degrader for ER+/HER2- Breast Cancer.
J.Med.Chem., 68, 2025
|
|
5V5F
 
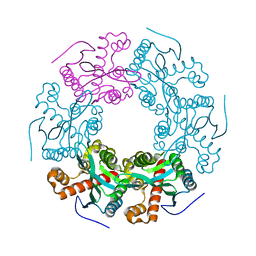 | | Crystal structure of RICE1 (PNT2) | | Descriptor: | At3g11770 | | Authors: | Li, P. | | Deposit date: | 2017-03-14 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.945 Å) | | Cite: | RISC-interacting clearing 3'- 5' exoribonucleases (RICEs) degrade uridylated cleavage fragments to maintain functional RISC in Arabidopsis thaliana.
Elife, 6, 2017
|
|
6K5H
 
 | |
5XJN
 
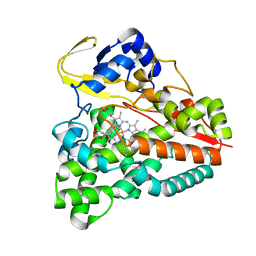 | | cytochrome P450 CREJ in complex with (4-ethylphenyl) dihydrogen phosphate | | Descriptor: | (4-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S, Feng, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6K5G
 
 | |
6K8P
 
 | |
6EFJ
 
 | | Crystal structure of NDM-1 with compound 9 | | Descriptor: | (2R)-2-phenyl-2-(phenylamino)-N-(1H-tetrazol-5-yl)acetamide, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Akhtar, A, Chen, Y. | | Deposit date: | 2018-08-16 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Active-Site Druggability of Carbapenemases and Broad-Spectrum Inhibitor Discovery.
Acs Infect Dis., 5, 2019
|
|
6K5K
 
 | |
7XLB
 
 | |
6K10
 
 | | Non substrate bound state of Staphylococcus Aureus AldH | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde dehydrogenase | | Authors: | Zhang, Z, Tao, X. | | Deposit date: | 2019-05-08 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.78962183 Å) | | Cite: | Structural Insight into the Substrate Gating Mechanism by Staphylococcus aureus Aldehyde Dehydrogenase
CCS Chemistry, 2, 2020
|
|
6KKH
 
 | | Crystal structure of the oxalate bound malyl-CoA lyase from Roseiflexus castenholzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase, MAGNESIUM ION, ... | | Authors: | Tang, W.R, Wang, Z.G, Zhang, C.Y, Wang, C. | | Deposit date: | 2019-07-25 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The C-terminal domain conformational switch revealed by the crystal structure of malyl-CoA lyase from Roseiflexus castenholzii.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6KIN
 
 | | Crystal structure of the tri-functional malyl-CoA lyase from Roseiflexus castenholzii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase | | Authors: | Tang, W.R, Zhang, C.Y, Wang, C, Xu, X.L. | | Deposit date: | 2019-07-19 | | Release date: | 2019-09-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.527 Å) | | Cite: | The C-terminal domain conformational switch revealed by the crystal structure of malyl-CoA lyase from Roseiflexus castenholzii.
Biochem.Biophys.Res.Commun., 518, 2019
|
|
6LJS
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-phenylphenyl)amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
7JRN
 
 | | Crystal structure of the wild type SARS-CoV-2 papain-like protease (PLPro) with inhibitor GRL0617 | | Descriptor: | 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, Non-structural protein 3, SULFATE ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-08-12 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay.
Acs Cent.Sci., 7, 2021
|
|
6LJX
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 2-phenylazanylbenzoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
4PAR
 
 | | The 5-Hydroxymethylcytosine-Specific Restriction Enzyme AbaSI in a Complex with Product-like DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA 14-MER, DNA 18-MER, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2014-04-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA.
Nucleic Acids Res., 42, 2014
|
|
6LJU
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[[3-chloranyl-4-(methylamino)-2-phenyl-phenyl]amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LJV
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 2-[[3-chloranyl-2-(2,3-dihydro-1-benzofuran-5-yl)phenyl]amino]benzoic acid, Fatty acid-binding protein, adipocyte | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6LQH
 
 | | High resolution architecture of curli complex | | Descriptor: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | Authors: | Zhang, M, Shi, H, Huang, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-15 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
6LQJ
 
 | | Low resolution architecture of curli complex | | Descriptor: | Curli production assembly/transport component CsgF, Curli production assembly/transport component CsgG | | Authors: | Zhang, M, Shi, H, Huang, Y. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Cryo-EM structure of the nonameric CsgG-CsgF complex and its implications for controlling curli biogenesis in Enterobacteriaceae.
Plos Biol., 18, 2020
|
|
9JAL
 
 | | Cryo-EM structure of MPXV core protease in complex with compound A1 | | Descriptor: | A1ECM-DI8-ALA-ETA, Core protease I7 | | Authors: | Gao, Y, Xie, X, Lan, W, Wang, W, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 643, 2025
|
|
9JAM
 
 | | Cryo-EM structure of MPXV core protease in complex with compound A3 | | Descriptor: | A1ECK-DI8-ALA-AEM, Core protease I7 | | Authors: | Gao, Y, Xie, X, Lan, W, Wang, W, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 643, 2025
|
|
9JAQ
 
 | | Cryo-EM structure of MPXV core protease in the apo-form | | Descriptor: | Core protease I7 | | Authors: | Lan, W, You, T, Li, D, Dong, X, Wang, H, Xu, J, Wang, W, Gao, Y, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 643, 2025
|
|
9JAN
 
 | | Cryo-EM structure of MPXV protease in complex with compound A4 | | Descriptor: | A1ECL-DI8-ALA-AEM, Core protease I7 | | Authors: | Gao, Y, Xie, X, Lan, W, Wang, W, Yang, H. | | Deposit date: | 2024-08-25 | | Release date: | 2025-03-12 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Substrate recognition and cleavage mechanism of the monkeypox virus core protease.
Nature, 643, 2025
|
|
6LJT
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3-chloranyl-2-phenyl-phenyl)amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
