5YPR
 
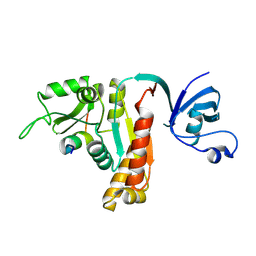 | | Crystal Structure of PSD-95 SH3-GK domain in complex with a synthesized inhibitor | | Descriptor: | Disks large homolog 4, Synthesized GK inhibitor | | Authors: | Zhu, J, Zhou, Q, Shang, Y, Weng, Z, Zhu, R, Zhang, M. | | Deposit date: | 2017-11-02 | | Release date: | 2018-03-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Synaptic Targeting and Function of SAPAPs Mediated by Phosphorylation-Dependent Binding to PSD-95 MAGUKs.
Cell Rep, 21, 2017
|
|
7UAK
 
 | |
5XS4
 
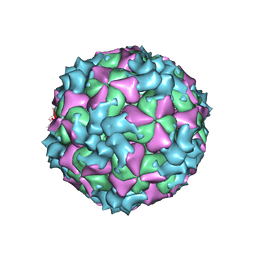 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS7
 
 | | Structure of Coxsackievirus A6 (CVA6) virus A-particle in complex with the neutralizing antibody fragment 1D5 | | Descriptor: | Genome polyprotein, Heavy chain of Fab 1D5, Light chain of Fab 1D5 | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Li, S.W, Cheng, T. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
5XS5
 
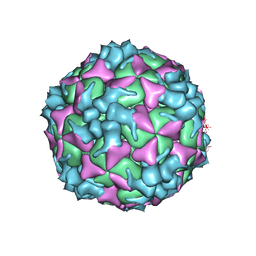 | | Structure of Coxsackievirus A6 (CVA6) virus procapsid particle | | Descriptor: | Genome polyprotein | | Authors: | Zheng, Q.B, He, M.Z, Xu, L.F, Yu, H, Cheng, T, Li, S.W. | | Deposit date: | 2017-06-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic structures of Coxsackievirus A6 and its complex with a neutralizing antibody
Nat Commun, 8, 2017
|
|
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-03-21 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7F0M
 
 | | Crystal Structure of human Pin1 complexed with a potent covalent inhibitor | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 8-(2-chloranylethanoyl)-4-[(5-naphthalen-1-ylfuran-2-yl)methyl]-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-06-05 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7F7G
 
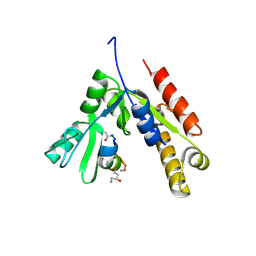 | | a linear Peptide Inhibitors in complex with GK domain | | Descriptor: | DLG4 GK domain, UNK-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYS-ALA-ILE-GLN-UNK | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
7DPF
 
 | | Cryo-EM structure of Coxsackievirus B1 mature virion | | Descriptor: | Capsid protein VP4, PALMITIC ACID, VP2, ... | | Authors: | Zheng, Q, Li, S. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures reveal the molecular basis of receptor-initiated coxsackievirus uncoating.
Cell Host Microbe, 29, 2021
|
|
7F7I
 
 | | Stapled Peptide Inhibitor in complex with PSD95 GK domain | | Descriptor: | ACE-ARG-ILE-ARG-ARG-ASP-GLU-TYR-LEU-LYZ-ALA-ILE-GLN-NH2, Disks large homolog 4 | | Authors: | Shang, Y, Huang, X, Li, X, Zhang, M. | | Deposit date: | 2021-06-29 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Entropy of stapled peptide inhibitors in free state is the major contributor to the improvement of binding affinity with the GK domain.
Rsc Chem Biol, 2, 2021
|
|
8BZI
 
 | | Human MST3 (STK24) kinase in complex with inhibitor MR39 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-azanylbutyl)-6-[2,5-bis(fluoranyl)-4-(6-methylpyridin-2-yl)phenyl]-2-(methylamino)pyrido[2,3-d]pyrimidin-7-one, Serine/threonine-protein kinase 24 | | Authors: | Balourdas, D.I, Rak, M, Tesch, R, Knapp, S, Joerger, A.C, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-12-14 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Shifting the selectivity of pyrido[2,3-d]pyrimidin-7(8H)-one inhibitors towards the salt-inducible kinase (SIK) subfamily.
Eur.J.Med.Chem., 254, 2023
|
|
7QRW
 
 | |
7QRX
 
 | |
