2CC9
 
 | | Complexes of Dodecin with Flavin and Flavin-like Ligands | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Grininger, M, Zeth, K, Oesterhelt, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-01-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dodecins: A Family of Lumichrome Binding Proteins.
J.Mol.Biol., 357, 2006
|
|
2CIF
 
 | | Complexes of Dodecin with Flavin and Flavin-like Ligands | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Grininger, M, Seiler, F, Zeth, K, Oesterhelt, D. | | Deposit date: | 2006-03-20 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dodecin Sequesters Fad in Closed Conformation from the Aqueous Solution.
J.Mol.Biol., 364, 2006
|
|
2CC7
 
 | | Complexes of Dodecin with Flavin and Flavin-like Ligands | | Descriptor: | CHLORIDE ION, LUMICHROME, MAGNESIUM ION, ... | | Authors: | Grininger, M, Zeth, K, Oesterhelt, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dodecins: A Family of Lumichrome Binding Proteins.
J.Mol.Biol., 357, 2006
|
|
2CIU
 
 | |
2WPQ
 
 | | Salmonella enterica SadA 479-519 fused to GCN4 adaptors (SadAK3, in- register fusion) | | Descriptor: | CHLORIDE ION, NITRATE ION, TRIMERIC AUTOTRANSPORTER ADHESIN FRAGMENT | | Authors: | Hartmann, M.D, Hernandez Alvarez, B, Albrecht, R, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-08-09 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Coiled-Coil Motif that Sequesters Ions to the Hydrophobic Core.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2XBP
 
 | | A novel signal transduction protein PII variant from Synechococcus elongatus PCC7942 indicates a two-step process for NAGK PII complex formation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Fokina, O, Chellamuthu, V.R, Zeth, K, Forchhammer, K. | | Deposit date: | 2010-04-14 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Novel Signal Transduction Protein P(II) Variant from Synechococcus Elongatus Pcc 7942 Indicates a Two-Step Process for Nagk- P(II) Complex Formation.
J.Mol.Biol., 399, 2010
|
|
2WG5
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4 | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2WA9
 
 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS - Trp peptide structure | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, TRP PEPTIDE | | Authors: | Schuenemann, V.J, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
2W1R
 
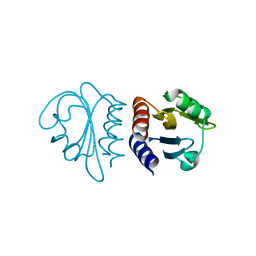 | | Crystal Structure of the C-terminal Domain of B. subtilis SpoVT | | Descriptor: | STAGE V SPORULATION PROTEIN T | | Authors: | Asen, I, Djuranovic, S, Lupas, A.N, Zeth, K. | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Spovt, the Final Modulator of Gene Expression During Spore Development in Bacillus Subtilis
J.Mol.Biol., 386, 2009
|
|
2WFW
 
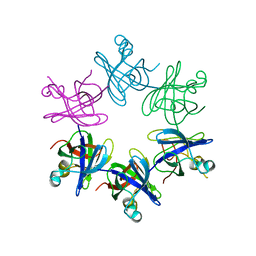 | | Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases - The Arc domain structure | | Descriptor: | ARC | | Authors: | Djuranovic, S, Hartmann, M.D, Habeck, M, Ursinus, A, Zwickl, P, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
2WA8
 
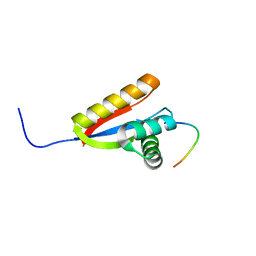 | | Structural basis of N-end rule substrate recognition in Escherichia coli by the ClpAP adaptor protein ClpS - The Phe peptide structure | | Descriptor: | ATP-DEPENDENT CLP PROTEASE ADAPTER PROTEIN CLPS, N-END RULE PEPTIDE | | Authors: | Schuenemann, V.J, Kralik, S.M, Albrecht, R, Spall, S.K, Truscott, K.N, Dougan, D.A, Zeth, K. | | Deposit date: | 2009-02-03 | | Release date: | 2009-04-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis of N-End Rule Substrate Recognition in Escherichia Coli by the Clpap Adaptor Protein Clps.
Embo Rep., 10, 2009
|
|
2X8X
 
 | |
2XTQ
 
 | |
2XTR
 
 | | Structure of the P176A Colicin M mutant from E. coli | | Descriptor: | COLICIN-M, NITRATE ION | | Authors: | Helbig, S, Patzer, S.I, Braun, V, Zeth, K. | | Deposit date: | 2010-10-12 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Activation of Colicin M by the Fkpa Prolyl Cis- Trans Isomerase/Chaperone.
J.Biol.Chem., 286, 2011
|
|
2YH3
 
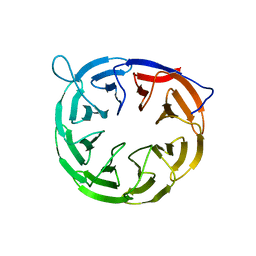 | | The structure of BamB from E. coli | | Descriptor: | LIPOPROTEIN YFGL | | Authors: | Albrecht, R, Zeth, K. | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis of Outer Membrane Protein Biogenesis in Bacteria.
J.Biol.Chem., 286, 2011
|
|
4GDO
 
 | | Structure of a fragment of the rod domain of plectin | | Descriptor: | Plectin | | Authors: | De Pereda, J.M, Buey, R.M, Uson, I, Sammito, M.D, De Marino, I. | | Deposit date: | 2012-08-01 | | Release date: | 2013-09-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploiting tertiary structure through local folds for crystallographic phasing.
Nat.Methods, 10, 2013
|
|
4RQY
 
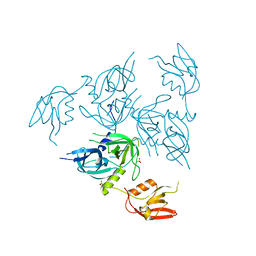 | |
2L7H
 
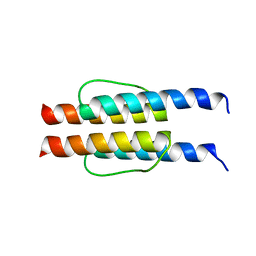 | |
2L7I
 
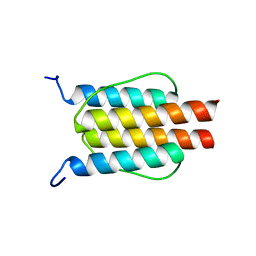 | |
3O2B
 
 | | E. coli ClpS in complex with a Phe N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, CHLORIDE ION, Phe N-end rule peptide, ... | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2H
 
 | | E. coli ClpS in complex with a Leu N-end rule peptide | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS, DNA protection during starvation protein | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
3O2O
 
 | | Structure of E. coli ClpS ring complex | | Descriptor: | ATP-dependent Clp protease adaptor protein ClpS | | Authors: | Roman-Hernandez, G, Grant, R.A, Sauer, R.T, Baker, T.A, de Regt, A. | | Deposit date: | 2010-07-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease.
Mol.Cell, 43, 2011
|
|
6V06
 
 | | Crystal structure of Beta-2 glycoprotein I purified from plasma (pB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-glycoprotein 1, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
6V09
 
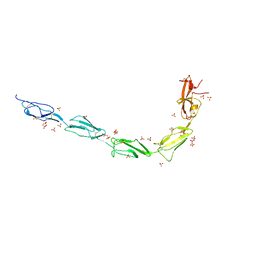 | | Crystal structure of human recombinant Beta-2 glycoprotein I short tag (ST-B2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
6V08
 
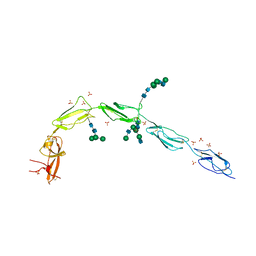 | | Crystal structure of human recombinant Beta-2 glycoprotein I (hrB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
