7TIY
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-48 | | Descriptor: | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-[(N-{[(2,4,5-trifluorophenyl)methoxy]carbonyl}-L-leucyl)amino]propane-1-sulfonic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIW
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB54 | | Descriptor: | (1S,2S)-2-[(N-{[(2-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIU
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB46 | | Descriptor: | (1S,2S)-2-[(N-{[(3-chlorophenyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIZ
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-63 | | Descriptor: | (1S,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-2-{[N-({[3-(trifluoromethyl)phenyl]methoxy}carbonyl)-L-leucyl]amino}propane-1-sulfonic acid, 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ... | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIA
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor NK01-14 | | Descriptor: | 3C-like proteinase nsp5, THIOCYANATE ION, benzyl [(2S)-3-cyclopropyl-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-1-oxopropan-2-yl]carbamate | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TIX
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor EB56 | | Descriptor: | 3C-like proteinase nsp5, MAGNESIUM ION, N~2~-{[(naphthalen-2-yl)methoxy]carbonyl}-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M.E, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
7TJ0
 
 | | Crystal structure of SARS-CoV-2 3CL in complex with inhibitor SL-4-241 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ACETATE ION | | Authors: | Forouhar, F, Liu, H, Iketani, S, Zack, A, Khanizeman, N, Bednarova, E, Fowler, B, Hong, S.J, Mohri, H, Nair, M.S, Huang, Y, Tay, N.E.S, Lee, S, Karan, C, Resnick, S.J, Quinn, C, Li, W, Shion, H, Jurtschenko, C, Lauber, M.A, McDonald, T, Stokes, M, Hurst, B, Rovis, T, Chavez, A, Ho, D.D, Stockwell, B.R. | | Deposit date: | 2022-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19.
Nat Commun, 13, 2022
|
|
4AYW
 
 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (PLATE FORM) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AYX
 
 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 (ROD FORM B) | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10, CARDIOLIPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AYT
 
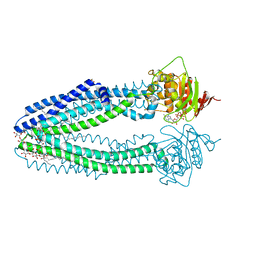 | | STRUCTURE OF THE HUMAN MITOCHONDRIAL ABC TRANSPORTER, ABCB10 | | Descriptor: | ATP-BINDING CASSETTE SUB-FAMILY B MEMBER 10 MITOCHONDRIAL, CARDIOLIPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Pike, A.C.W, Shintre, C.A, Li, Q, Kim, J, von Delft, F, Barr, A.J, Das, S, Chaikuad, A, Xia, X, Quigley, A, Dong, Y, Dong, L, Krojer, T, Vollmar, M, Muniz, J.R.C, Bray, J.E, Berridge, G, Chalk, R, Gileadi, O, Burgess-Brown, N, Shrestha, L, Goubin, S, Yang, J, Mahajan, P, Mukhopadhyay, S, Bullock, A.N, Arrowsmith, C.H, Weigelt, J, Bountra, C, Edwards, A.M, Carpenter, E.P. | | Deposit date: | 2012-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Abcb10, a Human ATP-Binding Cassette Transporter in Apo- and Nucleotide-Bound States
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4AW6
 
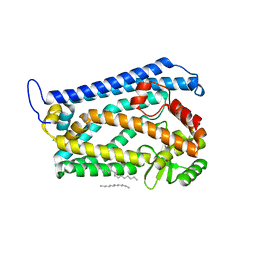 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 (FACE1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX PRENYL PROTEASE 1 HOMOLOG, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-05-31 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
2DD8
 
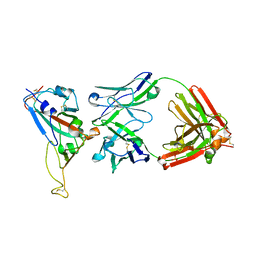 | | Crystal Structure of SARS-CoV Spike Receptor-Binding Domain Complexed with Neutralizing Antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IGG Heavy Chain, IGG Light Chain, ... | | Authors: | Prabakaran, P, Gan, J.H, Feng, Y, Zhu, Z.Y, Xiao, X.D, Ji, X, Dimitrov, D.S. | | Deposit date: | 2006-01-24 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Severe Acute Respiratory Syndrome Coronavirus Receptor-binding Domain Complexed with Neutralizing Antibody
J.Biol.Chem., 281, 2006
|
|
7WSB
 
 | | The ternary complex structure of FtmOx1 with a-ketoglutarate and 13-oxo-fumitremorgin B | | Descriptor: | 13-Oxofumitremorgin B, 2-OXOGLUTARIC ACID, COBALT (II) ION, ... | | Authors: | Wang, J, Wang, X.Y, Wang, Y.Y, Yan, W.P. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Dissecting the Mechanism of the Nonheme Iron Endoperoxidase FtmOx1 Using Substrate Analogues.
Jacs Au, 2, 2022
|
|
3ALA
 
 | | Crystal structure of vascular adhesion protein-1 in space group C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ernberg, K.E, McGrath, A.P, Guss, J.M. | | Deposit date: | 2010-07-29 | | Release date: | 2010-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A new crystal form of human vascular adhesion protein 1
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2G75
 
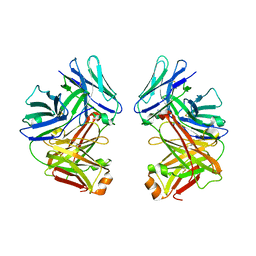 | | Crystal Structure of anti-SARS m396 Antibody | | Descriptor: | IGG Heavy Chain, IGG Light Chain | | Authors: | Prabakaran, P, Gan, J.H, Feng, Y, Zhu, Z.Y, Xiao, X.D, Ji, X, Dimitrov, D.S. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody.
J.Biol.Chem., 281, 2006
|
|
6GQF
 
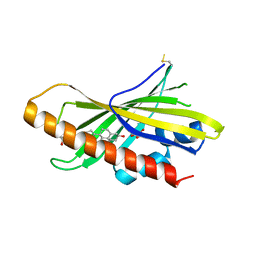 | | The structure of mouse AsterA (GramD1a) with 25-hydroxy cholesterol | | Descriptor: | 25-HYDROXYCHOLESTEROL, GLYCEROL, GRAM domain-containing protein 1A | | Authors: | Fairall, L, Gurnett, J.E, Vashi, D, Sandhu, J, Tontonoz, P, Schwabe, J.W.R. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Aster Proteins Facilitate Nonvesicular Plasma Membrane to ER Cholesterol Transport in Mammalian Cells.
Cell, 175, 2018
|
|
3DJA
 
 | | Crystal Structure of cpaf solved with MAD | | Descriptor: | Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-06-22 | | Release date: | 2009-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPN
 
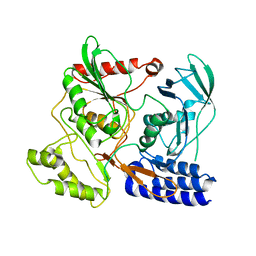 | | Crystal Structure of cpaf s499a mutant | | Descriptor: | Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DOR
 
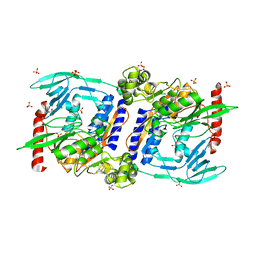 | | Crystal Structure of mature CPAF | | Descriptor: | Protein CT_858, SULFATE ION | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-06 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPM
 
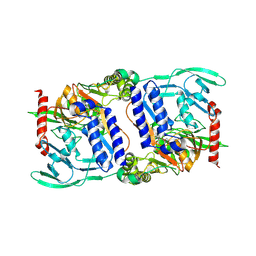 | | Structure of mature CPAF complexed with lactacystin | | Descriptor: | N-acetyl-S-({(2R,3S,4R)-3-hydroxy-2-[(1S)-1-hydroxy-2-methylpropyl]-4-methyl-5-oxopyrrolidin-2-yl}carbonyl)cysteine, Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
8FXF
 
 | | Crystal structure of the coiled-coil domain of TRIM56 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase TRIM56 | | Authors: | Lou, X.H, Ma, B.B, Zhuang, Y, Li, X.C. | | Deposit date: | 2023-01-24 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | TRIM56 coiled-coil domain structure provides insights into its E3 ligase functions.
Comput Struct Biotechnol J, 21, 2023
|
|
7Y42
 
 | |
7V97
 
 | | Arsenic-bound p53 DNA-binding domain mutant V272M | | Descriptor: | ARSENIC, Cellular tumor antigen p53, ZINC ION | | Authors: | Lu, M, Xing, Y.F, Wang, Z.Y, Ni, Y, Song, H.X. | | Deposit date: | 2021-08-24 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Diverse rescue potencies of p53 mutations to ATO are predetermined by intrinsic mutational properties.
Sci Transl Med, 15, 2023
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
7UG2
 
 | | Crystal structure of the coiled-coil domain of TRIM75 | | Descriptor: | ACETYL GROUP, ISOPROPYL ALCOHOL, Tripartite motif-containing protein 75 | | Authors: | Lou, X.H, Ma, B.B, Zhuang, Y, Li, X.C. | | Deposit date: | 2022-03-23 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Structural studies of the coiled-coil domain of TRIM75 reveal a tetramer architecture facilitating its E3 ligase complex.
Comput Struct Biotechnol J, 20, 2022
|
|
