4NUZ
 
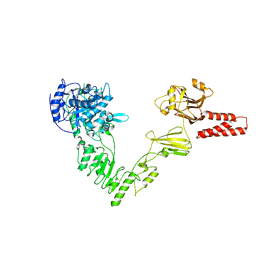 | | Crystal structure of a glycosynthase mutant (D233Q) of EndoS, an endo-beta-N-acetyl-glucosaminidase from Streptococcus pyogenes | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2 | | Authors: | Trastoy, B, Guenther, S, Snyder, G.A, Sundberg, E.J. | | Deposit date: | 2013-12-04 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NUY
 
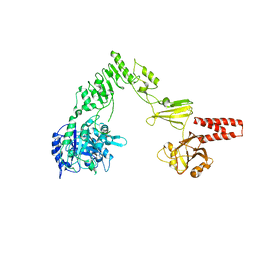 | | Crystal structure of EndoS, an endo-beta-N-acetyl-glucosaminidase from Streptococcus pyogenes | | Descriptor: | CALCIUM ION, Endo-beta-N-acetylglucosaminidase F2 | | Authors: | Trastoy, B, Guenther, S, Snyder, G.A, Sundberg, E.J. | | Deposit date: | 2013-12-04 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Crystal structure of Streptococcus pyogenes EndoS, an immunomodulatory endoglycosidase specific for human IgG antibodies.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4P0L
 
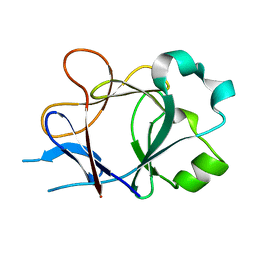 | |
4P0K
 
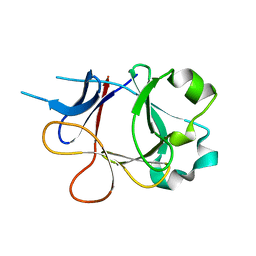 | |
6DHB
 
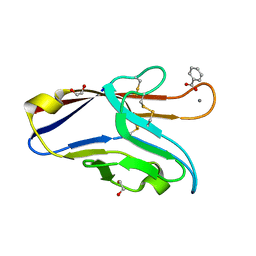 | | Crystal structure of the human TIM-3 with bound Calcium | | Descriptor: | 1,2-ETHANEDIOL, BENZOIC ACID, CALCIUM ION, ... | | Authors: | Gandhi, A.K, Kim, W.M, Huang, Y.H, Bonsor, D, Sundberg, E, Sun, Z.-Y, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2018-05-19 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High resolution X-ray and NMR structural study of human T-cell immunoglobulin and mucin domain containing protein-3.
Sci Rep, 8, 2018
|
|
5V91
 
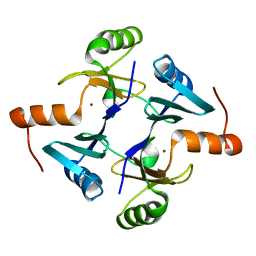 | | Crystal structure of fosfomycin resistance protein from Klebsiella pneumoniae | | Descriptor: | Fosfomycin resistance protein, ZINC ION | | Authors: | Klontz, E, Guenther, S, Silverstein, Z, Sundberg, E. | | Deposit date: | 2017-03-22 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and Dynamics of FosA-Mediated Fosfomycin Resistance in Klebsiella pneumoniae and Escherichia coli.
Antimicrob. Agents Chemother., 61, 2017
|
|
5V3D
 
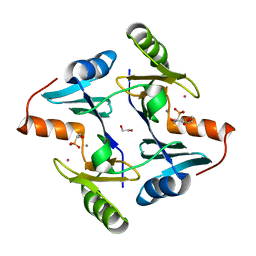 | | Crystal structure of fosfomycin resistance protein from Klebsiella pneumoniae with bound fosfomycin | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FOSFOMYCIN, ... | | Authors: | Klontz, E, Guenther, S, Silverstein, Z, Sundberg, E. | | Deposit date: | 2017-03-07 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.539 Å) | | Cite: | Structure and Dynamics of FosA-Mediated Fosfomycin Resistance in Klebsiella pneumoniae and Escherichia coli.
Antimicrob. Agents Chemother., 61, 2017
|
|
5VB0
 
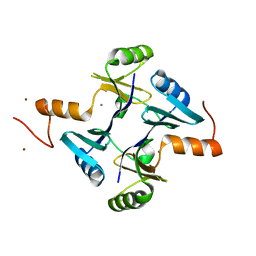 | | Crystal structure of fosfomycin resistance protein FosA3 | | Descriptor: | Fosfomycin resistance protein FosA3, MANGANESE (II) ION, NICKEL (II) ION | | Authors: | Klontz, E, Guenther, S, Silverstein, Z, Sundberg, E. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Structure and Dynamics of FosA-Mediated Fosfomycin Resistance in Klebsiella pneumoniae and Escherichia coli.
Antimicrob. Agents Chemother., 61, 2017
|
|
3BVG
 
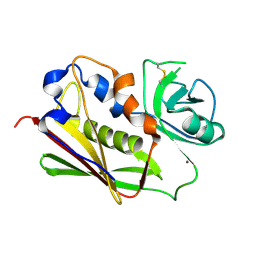 | |
3PL6
 
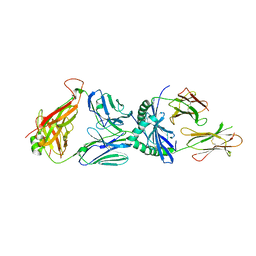 | |
6XNT
 
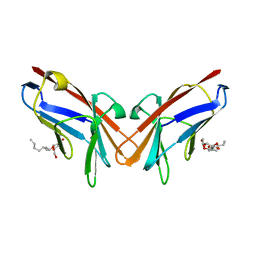 | | Crystal structure of I91A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, octyl beta-D-glucopyranoside | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
6XO1
 
 | | Crystal structure of N97A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-05 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.758 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
6XNW
 
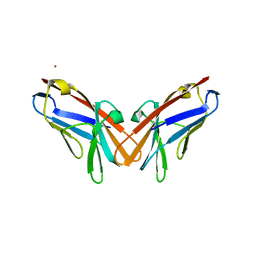 | | Crystal structure of V39A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, NICKEL (II) ION | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-04 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
6XNO
 
 | | Crystal structure of E99A mutant of human CEACAM1 | | Descriptor: | Carcinoembryonic antigen-related cell adhesion molecule 1, MALONIC ACID, octyl beta-D-glucopyranoside | | Authors: | Gandhi, A.K, Kim, W.M, Sun, Z.-Y, Huang, Y.H, Bonsor, D, Petsko, G.A, Kuchroo, V, Blumberg, R.S. | | Deposit date: | 2020-07-03 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the dynamic human CEACAM1 monomer-dimer equilibrium.
Commun Biol, 4, 2021
|
|
8F8V
 
 | | Crystal structure of Nb.X0 | | Descriptor: | Nb.X0 | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Andi, B. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
8F8W
 
 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, afucosylated IgG1 fragment | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
8F8X
 
 | | Crystal structure of Nb.X0 bound to the afucosylated human IgG1 fragment crystal form II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb.X0, Uncharacterized protein DKFZp686C11235 | | Authors: | Goldgur, Y, Ravetch, J, Gupta, A, Kao, K, Oren, D. | | Deposit date: | 2022-11-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism of glycoform specificity and in vivo protection by an anti-afucosylated IgG nanobody.
Nat Commun, 14, 2023
|
|
5DRZ
 
 | | Crystal structure of anti-HIV-1 antibody F240 Fab in complex with gp41 peptide | | Descriptor: | Envelope glycoprotein gp160, HIV Antibody F240 Heavy Chain, HIV Antibody F240 Light Chain, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2015-09-16 | | Release date: | 2016-10-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Molecular basis for epitope recognition by non-neutralizing anti-gp41 antibody F240.
Sci Rep, 6, 2016
|
|
5T87
 
 | | Crystal structure of CDI complex from Cupriavidus taiwanensis LMG 19424 | | Descriptor: | CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Target highlights from the first post-PSI CASP experiment (CASP12, May-August 2016).
Proteins, 86 Suppl 1, 2018
|
|
6V3P
 
 | |
5WJU
 
 | | Cryo-EM structure of B. subtilis flagellar filaments A39V, N133H | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WJY
 
 | |
5WJT
 
 | |
5WJZ
 
 | | Cryo-EM structure of B. subtilis flagellar filaments E115G | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
5WJW
 
 | | Cryo-EM structure of B. subtilis flagellar filaments H84R | | Descriptor: | Flagellin | | Authors: | Wang, F, Burrage, A.M, Orlova, A, Kearns, D.B, Egelman, E.H. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | A structural model of flagellar filament switching across multiple bacterial species.
Nat Commun, 8, 2017
|
|
