3E8Y
 
 | | Xray structure of scorpion toxin BmBKTx1 | | Descriptor: | CHLORIDE ION, Potassium channel toxin alpha-KTx 19.1, SULFATE ION | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-20 | | Release date: | 2009-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray structure of native scorpion toxin BmBKTx1 by racemic protein crystallography using direct methods.
J.Am.Chem.Soc., 131, 2009
|
|
3E7R
 
 | | X-ray Crystal Structure of Racemic Plectasin | | Descriptor: | Plectasin | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-18 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods
Protein Sci., 18, 2009
|
|
3E7U
 
 | | X-ray Crystal Structure of L-Plectasin | | Descriptor: | Plectasin | | Authors: | Mandal, K, Pentelute, B.L, Tereshko, V, Kossiakoff, A.A, Kent, S.B.H. | | Deposit date: | 2008-08-18 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods
Protein Sci., 18, 2009
|
|
5W2B
 
 | | Crystal structure of C-terminal domain of Ebola (Reston) nucleoprotein in complex with Fab fragment | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Nucleoprotein | | Authors: | Radwanska, M.J, Derewenda, U, Kossiakoff, A.A, Derewenda, Z.S. | | Deposit date: | 2017-06-06 | | Release date: | 2018-06-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of the C-terminal domain of the nucleoprotein from the Bundibugyo strain of the Ebola virus in complex with a pan-specific synthetic Fab.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8SLB
 
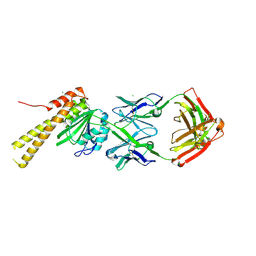 | | X-ray structure of CorA N-terminal domain in complex with conformation-specific synthetic antibody C12 | | Descriptor: | CHLORIDE ION, Cobalt/magnesium transport protein CorA, sAB C12 Heavy Chain, ... | | Authors: | Dominik, P.K, Erramilli, S.K, Reddy, B.G, Kossiakoff, A.A. | | Deposit date: | 2023-04-21 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Conformation-specific Synthetic Antibodies Discriminate Multiple Functional States of the Ion Channel CorA.
J.Mol.Biol., 435, 2023
|
|
4JQI
 
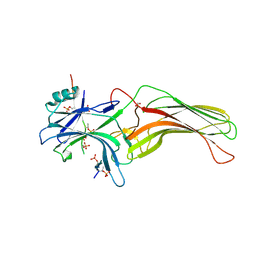 | | Structure of active beta-arrestin1 bound to a G protein-coupled receptor phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, Beta-arrestin-1, CHLORIDE ION, ... | | Authors: | Shukla, A.K, Manglik, A, Kruse, A.C, Xiao, K, Reis, R.I, Tseng, W.C, Staus, D.P, Hilger, D, Uysal, S, Huang, L.H, Paduch, M, Shukla, P.T, Koide, A, Koide, S, Weis, W.I, Kossiakoff, A.A, Kobilka, B.K, Lefkowitz, R.J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of active beta-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide.
Nature, 497, 2013
|
|
6CW2
 
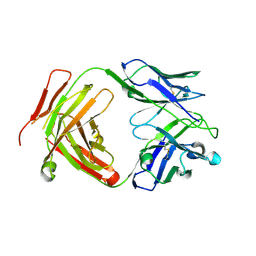 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 1 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CW3
 
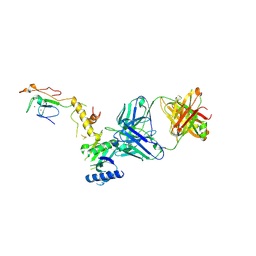 | | Crystal structure of a yeast SAGA transcriptional coactivator Ada2/Gcn5 HAT subcomplex, crystal form 2 | | Descriptor: | Histone acetyltransferase GCN5, Transcriptional adapter 2, ZINC ION, ... | | Authors: | Sun, J, Paduch, M, Kim, S.A, Kramer, R.M, Barrios, A.F, Lu, V, Luke, J, Usatyuk, S, Kossiakoff, A.A, Tan, S. | | Deposit date: | 2018-03-29 | | Release date: | 2018-09-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for activation of SAGA histone acetyltransferase Gcn5 by partner subunit Ada2.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CBV
 
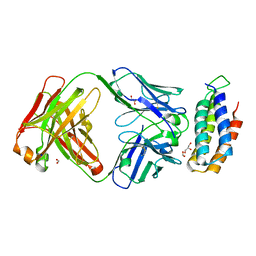 | | Crystal structure of BRIL bound to an affinity matured synthetic antibody. | | Descriptor: | BRIL, FORMIC ACID, GLYCEROL, ... | | Authors: | Mukherjee, S, Skrobek, B, Kossiakoff, A.A. | | Deposit date: | 2018-02-05 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
2FCS
 
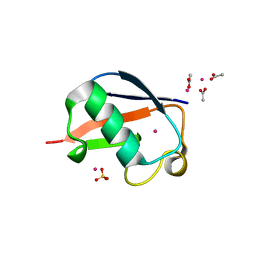 | | X-ray Crystal Structure of a Chemically Synthesized [L-Gln35]Ubiquitin with a Cubic Space Group | | Descriptor: | ACETATE ION, CADMIUM ION, SULFATE ION, ... | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FCN
 
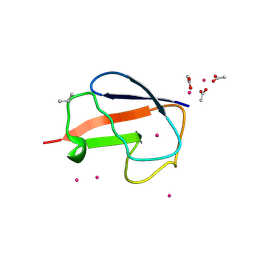 | | X-ray Crystal Structure of a Chemically Synthesized [D-Val35]Ubiquitin with a Cubic Space Group | | Descriptor: | ACETATE ION, CADMIUM ION, Ubiquitin | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FCQ
 
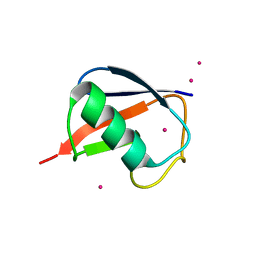 | | X-ray Crystal Structure of a Chemically Synthesized Ubiquitin with a Cubic Space Group | | Descriptor: | CADMIUM ION, Ubiquitin | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FD9
 
 | | X-ray Crystal Structure of Chemically Synthesized Crambin-{alpha}carboxamide | | Descriptor: | Crambin | | Authors: | Bang, D, Tereshko, V, Kossiakoff, A.A, Kent, S.B. | | Deposit date: | 2005-12-13 | | Release date: | 2007-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Role of a salt bridge in the model protein crambin explored by chemical protein synthesis: X-ray structure of a unique protein analogue, [V15A]crambin-alpha-carboxamide.
Mol Biosyst, 5, 2009
|
|
2FCM
 
 | | X-ray Crystal Structure of a Chemically Synthesized [D-Gln35]Ubiquitin with a Cubic Space Group | | Descriptor: | ACETATE ION, CADMIUM ION, Ubiquitin | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
3IVK
 
 | | Crystal Structure of the Catalytic Core of an RNA Polymerase Ribozyme Complexed with an Antigen Binding Antibody Fragment | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab heavy chain, ... | | Authors: | Koldobskaya, Y, Duguid, E.M, Shechner, D.M, Koide, S, Kossiakoff, A.A, Bartel, D.P, Piccirilli, J.A. | | Deposit date: | 2009-09-01 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
2FXU
 
 | | X-ray Structure of Bistramide A- Actin Complex at 1.35 A resolution. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Rizvi, S.A, Tereshko, V, Kossiakoff, A.A, Kozmin, S.A. | | Deposit date: | 2006-02-06 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of bistramide a-actin complex at a 1.35 A resolution
J.Am.Chem.Soc., 128, 2006
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
2FD7
 
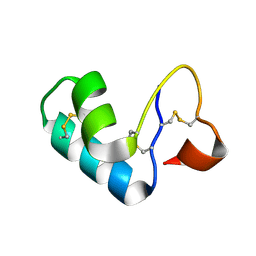 | | X-ray Crystal Structure of Chemically Synthesized Crambin | | Descriptor: | Crambin | | Authors: | Bang, D, Tereshko, V, Kossiakoff, A.A, Kent, S.B. | | Deposit date: | 2005-12-13 | | Release date: | 2007-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Role of a salt bridge in the model protein crambin explored by chemical protein synthesis: X-ray structure of a unique protein analogue, [V15A]crambin-alpha-carboxamide.
Mol Biosyst, 5, 2009
|
|
2HV2
 
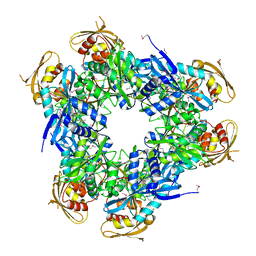 | | Crystal Structure of Conserved Protein of Unknown Function from Enterococcus faecalis V583 at 2.4 A Resolution, Probable N-Acyltransferase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Hypothetical protein, TETRAETHYLENE GLYCOL | | Authors: | Tereshko, V.A, Qiu, Y, Kossiakoff, A.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-07-27 | | Release date: | 2006-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of conserved hypothetical protein from Enterococcus faecalis V583 at 2.4 A resolution.
To be Published
|
|
7MJS
 
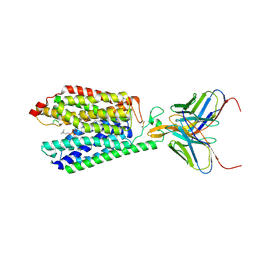 | | Single-Particle Cryo-EM Structure of Major Facilitator Superfamily Domain containing 2A in complex with LPC-18:3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2AG3 Fab heavy chain, 2AG3 Fab light chain, ... | | Authors: | Cater, R.J, Chua, G.L, Erramilli, S.K, Keener, J.E, Choy, B.C, Tokarz, P, Chin, C.F, Quek, D.Q.Y, Kloss, B, Pepe, J.G, Parisi, G, Wong, B.H, Clarke, O.B, Marty, M.T, Kossiakoff, A.A, Khelashvili, G, Silver, D.L, Mancia, F. | | Deposit date: | 2021-04-20 | | Release date: | 2021-06-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural basis of omega-3 fatty acid transport across the blood-brain barrier.
Nature, 595, 2021
|
|
7M74
 
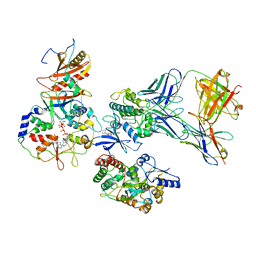 | | ATP-bound AMP-activated protein kinase | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Mukherjee, S, Harikumar, K.G, Strutzenberg, T, Zhou, X.E, Powell, S.K, Xu, T, Sheldon, R, Lamp, J, Brunzelle, J.S, Radziwon, K, Ellis, A, Novick, S.J, Vega, I.E, Jones, R, Miller, L.J, Xu, H.E, Griffin, P.R, Kossiakoff, A.A, Melcher, K. | | Deposit date: | 2021-03-26 | | Release date: | 2021-12-15 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structure of an AMPK complex in an inactive, ATP-bound state.
Science, 373, 2021
|
|
1AAP
 
 | | X-RAY CRYSTAL STRUCTURE OF THE PROTEASE INHIBITOR DOMAIN OF ALZHEIMER'S AMYLOID BETA-PROTEIN PRECURSOR | | Descriptor: | ALZHEIMER'S DISEASE AMYLOID A4 PROTEIN | | Authors: | Hynes, T.R, Randal, M, Kennedy, L.A, Eigenbrot, C, Kossiakoff, A.A. | | Deposit date: | 1990-09-14 | | Release date: | 1991-10-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of the protease inhibitor domain of Alzheimer's amyloid beta-protein precursor.
Biochemistry, 29, 1990
|
|
1BP3
 
 | | THE XRAY STRUCTURE OF A GROWTH HORMONE-PROLACTIN RECEPTOR COMPLEX | | Descriptor: | PROTEIN (GROWTH HORMONE), PROTEIN (PROLACTIN RECEPTOR), ZINC ION | | Authors: | Somers, W, Ultsch, M, De Vos, A.M, Kossiakoff, A.A. | | Deposit date: | 1998-08-12 | | Release date: | 1998-08-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The X-ray structure of a growth hormone-prolactin receptor complex.
Nature, 372, 1994
|
|
1D9C
 
 | | BOVINE INTERFERON-GAMMA AT 2.0 ANGSTROMS | | Descriptor: | INTERFERON-GAMMA | | Authors: | Randal, M, Kossiakoff, A.A. | | Deposit date: | 1999-10-27 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of bovine interferon-gamma; assessment of the structural differences between species.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1D9G
 
 | | BOVINE INTERFERON-GAMMA AT 2.9 ANGSTROMS | | Descriptor: | INTERFERON-GAMMA | | Authors: | Randal, M, Kossiakoff, A.A. | | Deposit date: | 1999-10-27 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The 2.0 A structure of bovine interferon-gamma; assessment of the structural differences between species.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
