4G0I
 
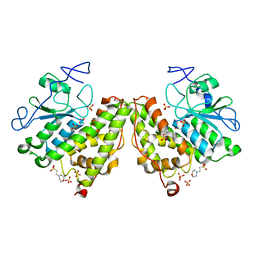 | | Glutathionyl-Hydroquinone Reductase, YqjG of Escherichia coli | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, protein yqjG | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
4FQU
 
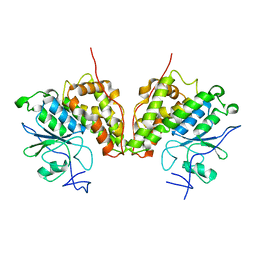 | |
4G0L
 
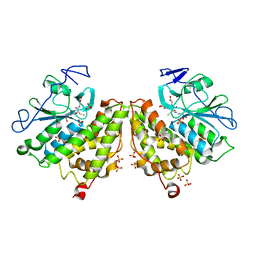 | | Glutathionyl-hydroquinone Reductase, YqjG, of E.coli complexed with GSH | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
4G5E
 
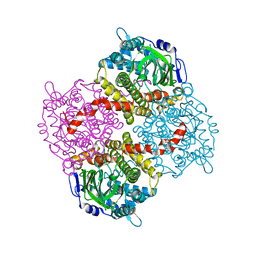 | | 2,4,6-Trichlorophenol 4-monooxygenase | | Descriptor: | 2,4,6-Trichlorophenol 4-monooxygenase | | Authors: | Hayes, R.P, Webb, B.N, Subramanian, A.K, Nissen, M, Popchock, A, Xun, L, Kang, C. | | Deposit date: | 2012-07-17 | | Release date: | 2012-09-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Catalytic Differences between Two FADH(2)-Dependent Monooxygenases: 2,4,5-TCP 4-Monooxygenase (TftD) from Burkholderia cepacia AC1100 and 2,4,6-TCP 4-Monooxygenase (TcpA) from Cupriavidus necator JMP134.
Int J Mol Sci, 13, 2012
|
|
4G0K
 
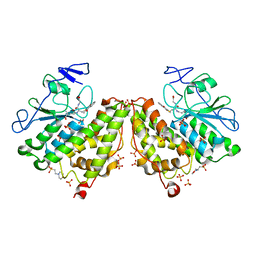 | | Glutathionyl-hydroquinone reductase, YqjG, of E.coli complexed with GS-menadione | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, L-gamma-glutamyl-S-(3-methyl-1,4-dioxo-1,4-dihydronaphthalen-2-yl)-L-cysteinylglycine, SULFATE ION, ... | | Authors: | Green, A.R, Hayes, R.P, Xun, L, Kang, C. | | Deposit date: | 2012-07-09 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.557 Å) | | Cite: | Structural understanding of the glutathione-dependent reduction mechanism of glutathionyl-hydroquinone reductases.
J.Biol.Chem., 287, 2012
|
|
2J3J
 
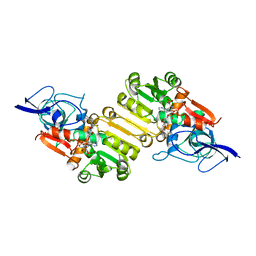 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex I | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2J3K
 
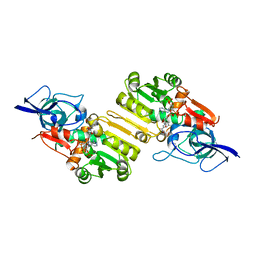 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex II | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2J3I
 
 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Binary Complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT OXIDOREDUCTASE P1 | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and Structural Studies of Apoform, Binary, and Ternary Complexes of the Arabidopsis Alkenal Double Bond Reductase at5G16970.
J.Biol.Chem., 281, 2006
|
|
6AFK
 
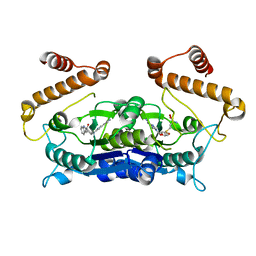 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-{(3S)-1-[3-(pyridin-4-yl)-1H-pyrazol-5-yl]piperidin-3-yl}-1H-indole-2-carboxamide, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Koay, A, Wong, Y.W, Sahili, A.E, Nah, Q, Kang, C, Poulsen, A, Chionh, Y.K, McBee, M, Matter, A, Hill, J, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Targeting the Bacterial Epitranscriptome for Antibiotic Development: Discovery of Novel tRNA-(N1G37) Methyltransferase (TrmD) Inhibitors.
Acs Infect Dis., 5, 2019
|
|
5ZMA
 
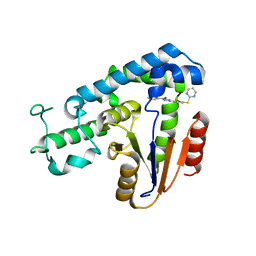 | | Structural basis for an allosteric Eya2 phosphatase inhibitor | | Descriptor: | 3-fluoro-N'-[(E)-{5-[(pyrimidin-2-yl)sulfanyl]furan-2-yl}methylidene]benzohydrazide, Eyes absent homolog 2 | | Authors: | Anantharajan, J, Jansson, A.E, Kang, C. | | Deposit date: | 2018-04-02 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.175 Å) | | Cite: | Structural and Functional Analyses of an Allosteric EYA2 Phosphatase Inhibitor That Has On-Target Effects in Human Lung Cancer Cells.
Mol.Cancer Ther., 18, 2019
|
|
6AT7
 
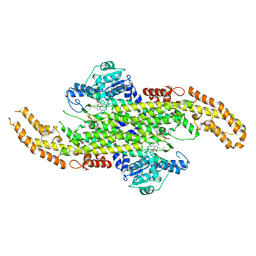 | | Phenylalanine Ammonia-Lyase (PAL) from Sorghum bicolor | | Descriptor: | AMMONIUM ION, Phenylalanine ammonia-lyase | | Authors: | Jun, S.Y, Kang, C. | | Deposit date: | 2017-08-28 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.493 Å) | | Cite: | Biochemical and Structural Analysis of Substrate Specificity of a Phenylalanine Ammonia-Lyase.
Plant Physiol., 176, 2018
|
|
2LAT
 
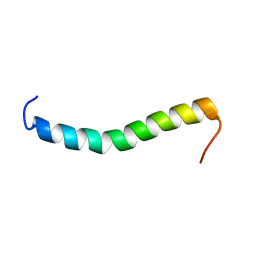 | | Solution structure of a Human minimembrane protein OST4 | | Descriptor: | Dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 4 | | Authors: | Gayen, S, Kang, C. | | Deposit date: | 2011-03-21 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a human minimembrane protein Ost4, a subunit of the oligosaccharyltransferase complex.
Biochem.Biophys.Res.Commun., 409, 2011
|
|
2M0S
 
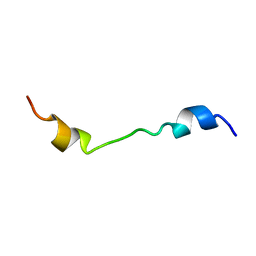 | |
2IUK
 
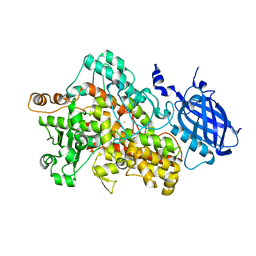 | | Crystal structure of Soybean Lipoxygenase-D | | Descriptor: | FE (III) ION, SEED LIPOXYGENASE | | Authors: | Youn, B, Sellhorn, G.E, Mirchel, R.J, Gaffney, B.J, Grimes, H.D, Kang, C. | | Deposit date: | 2006-06-05 | | Release date: | 2006-10-11 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Vegetative Soybean Lipoxygenase Vlx-B and Vlx-D, and Comparisons with Seed Isoforms Lox-1 and Lox-3.
Proteins, 65, 2006
|
|
2IUJ
 
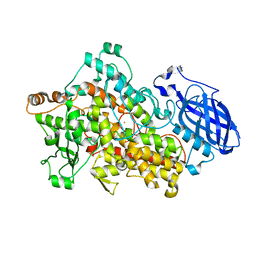 | | Crystal Structure of Soybean Lipoxygenase-B | | Descriptor: | FE (III) ION, LIPOXYGENASE L-5 | | Authors: | Youn, B, Sellhorn, G.E, Mirchel, R.J, Gaffney, B.J, Grimes, H.D, Kang, C. | | Deposit date: | 2006-06-05 | | Release date: | 2006-10-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Vegetative Soybean Lipoxygenase Vlx-B and Vlx-D, and Comparisons with Seed Isoforms Lox-1 and Lox-3.
Proteins, 65, 2006
|
|
2MFR
 
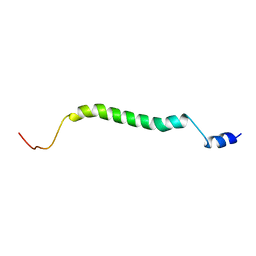 | |
2MV6
 
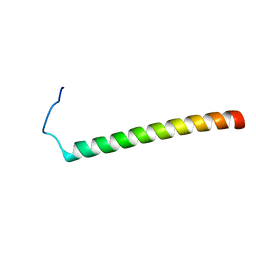 | |
2N7G
 
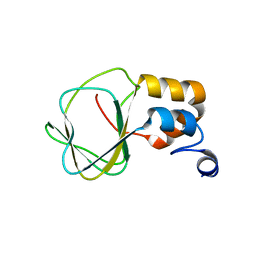 | |
2MXB
 
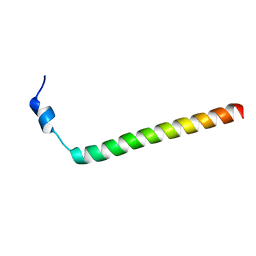 | |
2N7Q
 
 | |
2N7R
 
 | |
2L4R
 
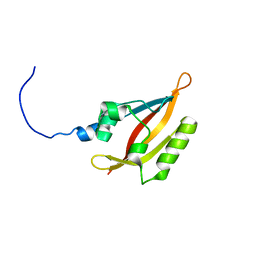 | | NMR solution structure of the N-terminal PAS domain of hERG | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Gayen, N, Li, Q, Chen, A.S, Huang, Q, Raida, M, Kang, C. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the N-terminal domain of hERG and its interaction with the S4-S5 linker.
Biochem.Biophys.Res.Commun., 403, 2010
|
|
2NDJ
 
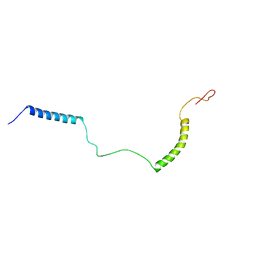 | | Structural Basis for KCNE3 and Estrogen Modulation of the KCNQ1 Channel | | Descriptor: | Potassium voltage-gated channel subfamily E member 3 | | Authors: | Sanders, C.R, Van Horn, W.D, Kroncke, B.M, Sisco, N.J, Meiler, J, Vanoye, C.G, Song, Y, Nannemann, D.P, Welch, R.C, Kang, C, Smith, J, George, A.L. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for KCNE3 modulation of potassium recycling in epithelia.
Sci Adv, 2, 2016
|
|
8FEM
 
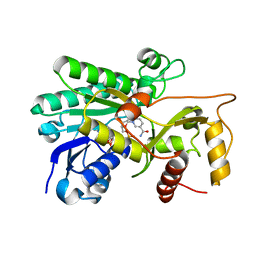 | | Panicum vigratum Dihydroflavonol 4-reductase complexed with NADP | | Descriptor: | Dihydroflavonol 4-Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lewis, J.A, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
8FET
 
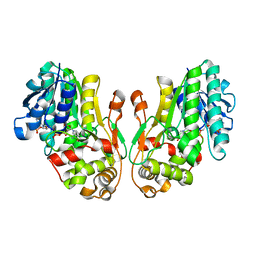 | | Flavanone 4-Reductase from Sorghum bicolor-NADP(H) complex | | Descriptor: | 3-deoxyanthocyanidin synthase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-12-06 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural Similarities and Overlapping Activities among Dihydroflavonol 4-Reductase, Flavanone 4-Reductase, and Anthocyanidin Reductase Offer Metabolic Flexibility in the Flavonoid Pathway.
Int J Mol Sci, 24, 2023
|
|
