3AJ2
 
 | | The structure of AxCeSD octamer (C-terminal HIS-tag) from Acetobacter xylinum | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Tanaka, I, Yao, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3AJ1
 
 | | The structure of AxCeSD octamer (N-terminal HIS-tag) from Acetobacter xylinum | | Descriptor: | Cellulose synthase operon protein D | | Authors: | Hu, S.Q, Tajima, K, Zhou, Y, Tanaka, I, Yao, M. | | Deposit date: | 2010-05-20 | | Release date: | 2010-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial cellulose synthase subunit D octamer with four inner passageways
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4M3Q
 
 | | Crystal structure of the catalytic domain of the proto-oncogene tyrosine-protein kinase MER in complex with inhibitor UNC1917 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Tyrosine-protein kinase Mer, ... | | Authors: | Zhang, W, Zhang, D, Stashko, M.A, DeRyckere, D, Hunter, D, Kireev, D.B, Miley, M, Cummings, C, Lee, M, Norris-Drouin, J, Stewart, W.M, Sather, S, Zhou, Y, Kirkpatrick, G, Machius, M, Janzen, W.P, Earp, H.S, Graham, D.K, Frye, S, Wang, X. | | Deposit date: | 2013-08-06 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.718 Å) | | Cite: | Pseudo-Cyclization through Intramolecular Hydrogen Bond Enables Discovery of Pyridine Substituted Pyrimidines as New Mer Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
2DGN
 
 | | Mouse Muscle Adenylosuccinate Synthetase partially ligated complex with GTP, 2'-deoxy-IMP | | Descriptor: | 9-(2-DEOXY-5-O-PHOSPHONO-BETA-D-ERYTHRO-PENTOFURANOSYL)-6-(PHOSPHONOOXY)-9H-PURINE, Adenylosuccinate synthetase isozyme 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Iancu, C.V, Zhou, Y, Borza, T, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2006-03-15 | | Release date: | 2006-09-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cavitation as a mechanism of substrate discrimination by adenylosuccinate synthetases.
Biochemistry, 45, 2006
|
|
8YP3
 
 | | Crystal structure of UDP-N-acetylglucosamine pyrophosphorylase from Spodoptera frugiperda in complex with UDP-GlcNAc | | Descriptor: | MAGNESIUM ION, SULFATE ION, UDP-N-acetylglucosamine diphosphorylase, ... | | Authors: | Lu, Q, Liu, T, Zhou, Y, Yang, Q. | | Deposit date: | 2024-03-15 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and Inhibition of Insect UDP- N -acetylglucosamine Pyrophosphorylase: A Key Enzyme in the Hexosamine Biosynthesis Pathway.
J.Agric.Food Chem., 72, 2024
|
|
4V60
 
 | | The structure of rat liver vault at 3.5 angstrom resolution | | Descriptor: | Major vault protein | | Authors: | Kato, K, Zhou, Y, Tanaka, H, Yao, M, Yamashita, E, Yoshimura, M, Tsukihara, T. | | Deposit date: | 2008-10-24 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of rat liver vault at 3.5 angstrom resolution
Science, 323, 2009
|
|
6FCE
 
 | | NMR ensemble of Macrocyclic Peptidomimetic Containing Constrained a,a-dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors | | Descriptor: | ACP-HIS-DPHE-ARG-TRP-ASP-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Grieco, P, Merlino, F, Zhou, Y, Cai, M, Yousif, A.M, Di Maro, S, Novellino, E, Hruby, V.J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Development of Macrocyclic Peptidomimetics Containing Constrained alpha , alpha-Dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors.
J. Med. Chem., 61, 2018
|
|
6ZUQ
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | Extracellular protein 11-1, GLYCEROL, ZINC ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2022-07-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
6ZUS
 
 | | Crystal structure of the effector Ecp11-1 from Fulvia fulva | | Descriptor: | DI(HYDROXYETHYL)ETHER, Extracellular protein 11-1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-07-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
7AD5
 
 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | Descriptor: | ACETATE ION, Avirulence protein LmJ1, GLYCEROL, ... | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-09-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
7B76
 
 | | Crystal structure of the effector AvrLm5-9 from Leptosphaeria maculans | | Descriptor: | Avirulence protein LmJ1, IODIDE ION | | Authors: | Lazar, N, Mesarich, C, Petit-Houdenot, Y, Talbi, N, Li de la Sierra-Gallay, I, Zelie, E, Blondeau, K, Gracy, J, Ollivier, B, van de Wouw, A, Balesdent, M.H, Idnurm, A, van Tilbeurgh, H, Fudal, I. | | Deposit date: | 2020-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new family of structurally conserved fungal effectors displays epistatic interactions with plant resistance proteins.
Plos Pathog., 18, 2022
|
|
6GHM
 
 | | Structure of PP1 alpha phosphatase bound to ASPP2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Apoptosis-stimulating of p53 protein 2, ... | | Authors: | Mouilleron, S, Bertran, T.M, Tapon, N, Zhou, Y. | | Deposit date: | 2018-05-08 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | ASPP proteins discriminate between PP1 catalytic subunits through their SH3 domain and the PP1 C-tail.
Nat Commun, 10, 2019
|
|
8T2I
 
 | | Negative stain EM assembly of MYC, JAZ, and NINJA complex | | Descriptor: | AFP homolog 2, Maltose/maltodextrin-binding periplasmic protein, Protein TIFY 10A, ... | | Authors: | Zhou, X.E, Zhang, Y, Zhou, Y, He, Q, Cao, X, Kariapper, L, Suino-Powell, K, Zhu, Y, Zhang, F, Karsten, M. | | Deposit date: | 2023-06-06 | | Release date: | 2023-06-28 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (10.4 Å) | | Cite: | Assembly of JAZ-JAZ and JAZ-NINJA complexes in jasmonate signaling.
Plant Commun., 4, 2023
|
|
7QA9
 
 | | 10bp DNA/DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*TP*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
8XP1
 
 | | Cryo-EM structure of the protomers of the C ring in the CW state | | Descriptor: | Chemotaxis protein CheY, Flagellar M-ring protein, Flagellar motor switch protein FliG, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the protomers of the C ring in the CW state
To Be Published
|
|
8XP0
 
 | | Cryo-EM structure of the protomers of the C ring in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the protomers of the C ring in the CCW state
To Be Published
|
|
8YJT
 
 | | Cryo-EM structure of the flagellar C ring in the CCW state | | Descriptor: | Flagellar M-ring protein, Flagellar motor switch protein FliG, Flagellar motor switch protein FliM, ... | | Authors: | Tan, J.X, Zhang, L, Zhou, Y, Zhu, Y.Q. | | Deposit date: | 2024-03-02 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structure of the flagellar C ring in the CCW state
To Be Published
|
|
6XDG
 
 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies | | Descriptor: | REGN10933 antibody Fab fragment heavy chain, REGN10933 antibody Fab fragment light chain, REGN10987 antibody Fab fragment heavy chain, ... | | Authors: | Franklin, M.C, Saotome, K, Romero Hernandez, A, Zhou, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-06-24 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail.
Science, 369, 2020
|
|
6L15
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L12
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 4-[(2-chloranylphenoxazin-10-yl)methyl]cyclohexan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6S47
 
 | | Saccharomyces cerevisiae 80S ribosome bound with ABCF protein New1 | | Descriptor: | 18S rRNA (1707-MER), 28S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Kasari, V, Pochopien, A.A, Margus, T, Murina, V, Turnbull, K, Zhou, Y, Nissan, T, Graf, M, Novacek, J, Atkinson, G.C, Johansson, M.J.O, Wilson, D.N, Hauryliuk, V. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-24 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A role for the Saccharomyces cerevisiae ABCF protein New1 in translation termination/recycling.
Nucleic Acids Res., 47, 2019
|
|
6L16
 
 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-[4-[2-(7-chloranylpyrido[3,4-b][1,4]benzoxazin-5-yl)ethyl]piperidin-1-yl]ethanamine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L11
 
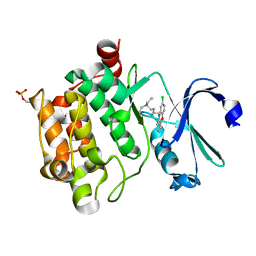 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 5-(2-chloranylphenoxazin-10-yl)-~{N},~{N}-diethyl-pentan-1-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L17
 
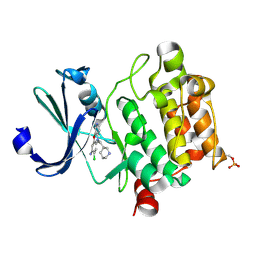 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 7-chloranyl-5-[3-[(3~{S})-piperidin-3-yl]propyl]pyrido[3,4-b][1,4]benzoxazin-8-amine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6L13
 
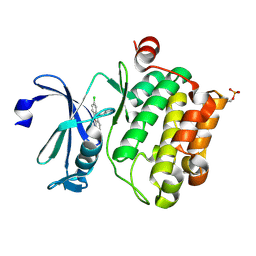 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-(2-piperidin-4-ylethyl)phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
