8D7H
 
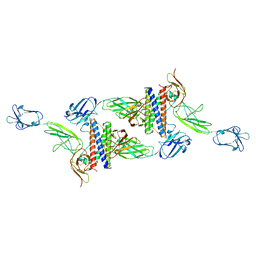 | | Cryo-EM structure of human CLCF1 in complex with CRLF1 and CNTFR alpha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cardiotrophin-like cytokine factor 1, ... | | Authors: | Zhou, Y, Franklin, M.C. | | Deposit date: | 2022-06-07 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into the assembly of gp130 family cytokine signaling complexes.
Sci Adv, 9, 2023
|
|
8D82
 
 | |
8D85
 
 | |
8ESB
 
 | |
8ESA
 
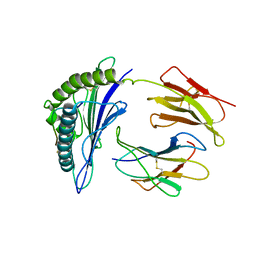 | |
8ES9
 
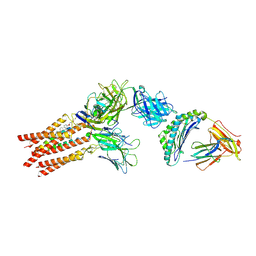 | | CryoEM structure of PN45428 TCR-CD3 in complex with HLA-A2 MAGEA4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Saotome, K, Franklin, M.C. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM.
Nat Commun, 14, 2023
|
|
8ES7
 
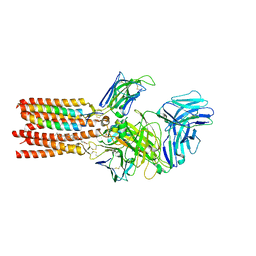 | | CryoEM structure of PN45545 TCR-CD3 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Saotome, K, Franklin, M.C. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural analysis of cancer-relevant TCR-CD3 and peptide-MHC complexes by cryoEM.
Nat Commun, 14, 2023
|
|
8ES8
 
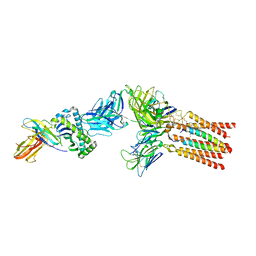 | |
3TRJ
 
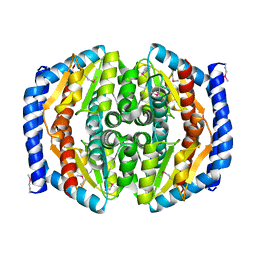 | | Structure of a phosphoheptose isomerase from Francisella tularensis | | Descriptor: | Phosphoheptose isomerase | | Authors: | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rapid countermeasure discovery against Francisella tularensis based on a metabolic network reconstruction.
Plos One, 8, 2013
|
|
5HF9
 
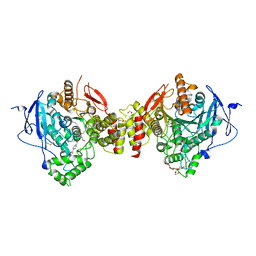 | | Crystal structure of human acetylcholinesterase in complex with paraoxon and HI6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Franklin, M.F, Rudolph, M.J, Ginter, C, Cassidy, M.S, Cheung, J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of paraoxon-inhibited human acetylcholinesterase reveal perturbations of the acyl loop and the dimer interface.
Proteins, 84, 2016
|
|
5HF8
 
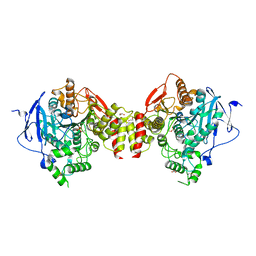 | | Crystal structure of human acetylcholinesterase in complex with paraoxon (alternative acyl loop conformation) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Franklin, M.F, Rudolph, M.J, Ginter, C, Cassidy, M.S, Cheung, J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of paraoxon-inhibited human acetylcholinesterase reveal perturbations of the acyl loop and the dimer interface.
Proteins, 84, 2016
|
|
5HF5
 
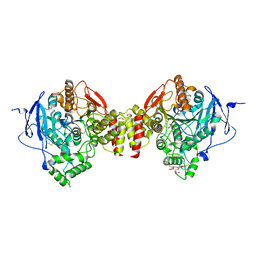 | | Crystal structure of human acetylcholinesterase in complex with paraoxon in the unaged state (predominant acyl loop conformation) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Franklin, M.F, Rudolph, M.J, Ginter, C, Cassidy, M.S, Cheung, J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Structures of paraoxon-inhibited human acetylcholinesterase reveal perturbations of the acyl loop and the dimer interface.
Proteins, 84, 2016
|
|
5HF6
 
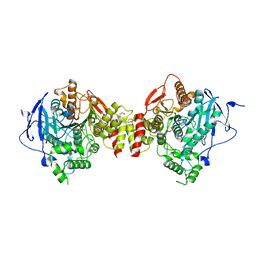 | | Crystal structure of human acetylcholinesterase in complex with paraoxon in the aged state | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Franklin, M.F, Rudolph, M.J, Ginter, C, Cassidy, M.S, Cheung, J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of paraoxon-inhibited human acetylcholinesterase reveal perturbations of the acyl loop and the dimer interface.
Proteins, 84, 2016
|
|
5HFA
 
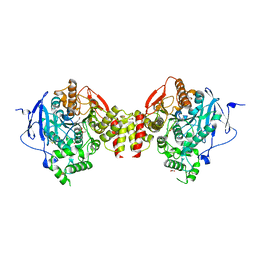 | | Crystal structure of human acetylcholinesterase in complex with paraoxon and 2-PAM | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Franklin, M.F, Rudolph, M.J, Ginter, C, Cassidy, M.S, Cheung, J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-06-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of paraoxon-inhibited human acetylcholinesterase reveal perturbations of the acyl loop and the dimer interface.
Proteins, 84, 2016
|
|
6XDG
 
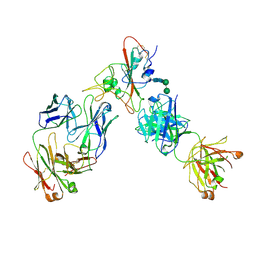 | | Complex of SARS-CoV-2 receptor binding domain with the Fab fragments of two neutralizing antibodies | | Descriptor: | REGN10933 antibody Fab fragment heavy chain, REGN10933 antibody Fab fragment light chain, REGN10987 antibody Fab fragment heavy chain, ... | | Authors: | Franklin, M.C, Saotome, K, Romero Hernandez, A, Zhou, Y. | | Deposit date: | 2020-06-10 | | Release date: | 2020-06-24 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail.
Science, 369, 2020
|
|
8EPA
 
 | |
3TR2
 
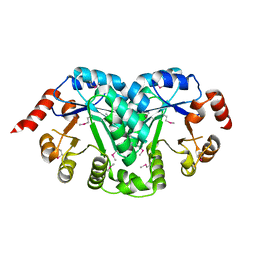 | | Structure of a orotidine 5'-phosphate decarboxylase (pyrF) from Coxiella burnetii | | Descriptor: | Orotidine 5'-phosphate decarboxylase | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TRF
 
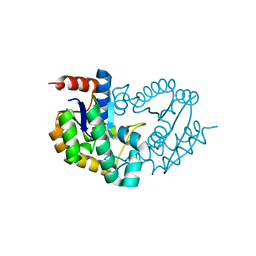 | | Structure of a shikimate kinase (aroK) from Coxiella burnetii | | Descriptor: | SULFATE ION, Shikimate kinase | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TR0
 
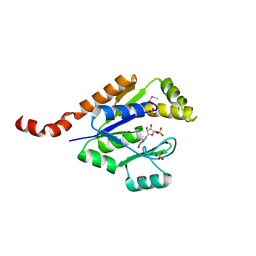 | | Structure of guanylate kinase (gmk) from Coxiella burnetii | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Guanylate kinase, SULFATE ION | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3UWC
 
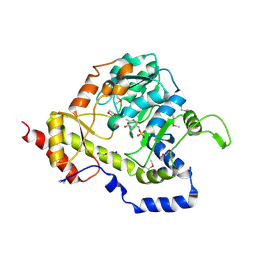 | | Structure of an aminotransferase (DegT-DnrJ-EryC1-StrS family) from Coxiella burnetii in complex with PMP | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-12-01 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TQZ
 
 | | Structure of a deoxyuridine 5'-triphosphate nucleotidohydrolase (dut) from Coxiella burnetii | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, SULFATE ION | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
4IEG
 
 | |
5JI3
 
 | | HslUV complex | | Descriptor: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
5JI2
 
 | | HslU L199Q in HslUV complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent protease ATPase subunit HslU, ATP-dependent protease subunit HslV, ... | | Authors: | Grant, R.A, Sauer, R.T, Schmitz, K.R, Baytshtok, V. | | Deposit date: | 2016-04-21 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.307 Å) | | Cite: | A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Structure, 24, 2016
|
|
3CFP
 
 | | Structure of the replicating complex of a POL Alpha family DNA Polymerase, ternary complex 1 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*DAP*DCP*DAP*DGP*DGP*DTP*DAP*DAP*DGP*DCP*DAP*DGP*DTP*DCP*DCP*DGP*DCP*DG)-3'), ... | | Authors: | Wang, J, Klimenko, D, Wang, M, Steitz, T.A, Konigsberg, W.H. | | Deposit date: | 2008-03-04 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into base selectivity from the structures
of an RB69 DNA Polymerase triple mutant
To be Published
|
|
