5IJB
 
 | | The ligand-free structure of the mouse TLR4/MD-2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 96, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5IJD
 
 | | The crystal structure of mouse TLR4/MD-2/lipid A complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, Y, Su, L, Morin, M.D, Jones, B.T, Whitby, L.R, Surakattula, M, Huang, H, Shi, H, Choi, J.H, Wang, K, Moresco, E.M, Berger, M, Zhan, X, Zhang, H, Boger, D.L, Beutler, B. | | Deposit date: | 2016-03-01 | | Release date: | 2016-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3ZYH
 
 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALBG0 AT 1.5 A RESOLUTION | | Descriptor: | 3-(beta-D-galactopyranosylthio)propanoic acid, CALCIUM ION, PA-I galactophilic lectin | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3ZYB
 
 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALAG0 AT 2.3 A RESOLUTION | | Descriptor: | CALCIUM ION, GALA-LYS-PRO-LEUNH2, P-HYDROXYBENZOIC ACID, ... | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-19 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3ZYF
 
 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH NPG AT 1.9 A RESOLUTION | | Descriptor: | 4-nitrophenyl beta-D-galactopyranoside, CALCIUM ION, PA-I GALACTOPHILIC LECTIN | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-22 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
8QOY
 
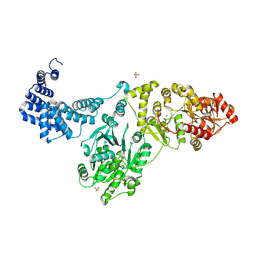 | | Capsular polysaccharide synthesis multienzyme of Actinobacillus Pleuropneumoniae serotype 3 | | Descriptor: | SULFATE ION, TagF-like capsule polymerase Cps3D, ZINC ION | | Authors: | Di Domenico, V, Litschko, C, Schulze, J, Bethe, A, Cifuente, J.O, Marina, A, Budde, I, Mast, T.A, Sulewska, M, Berger, M, Buettner, F, Gerardy-Schahn, R, Fiebig, T, Guerin, M.E. | | Deposit date: | 2023-09-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Transition transferases prime bacterial capsule polymerization.
Nat.Chem.Biol., 2024
|
|
1MVR
 
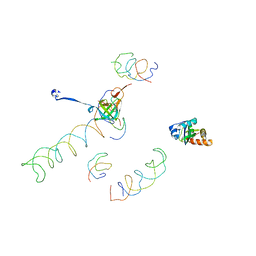 | | Decoding Center & Peptidyl transferase center from the X-ray structure of the Thermus thermophilus 70S ribosome, aligned to the low resolution Cryo-EM map of E.coli 70S Ribosome | | Descriptor: | 30S RIBOSOMAL PROTEIN S12, 50S ribosomal protein L11, Helix 34 of 16S rRNA, ... | | Authors: | Rawat, U.B, Zavialov, A.V, Sengupta, J, Valle, M, Grassucci, R.A, Linde, J, Vestergaard, B, Ehrenberg, M, Frank, J. | | Deposit date: | 2002-09-26 | | Release date: | 2003-04-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | A cryo-electron microscopic study of ribosome-bound termination factor RF2
Nature, 421, 2003
|
|
1PN8
 
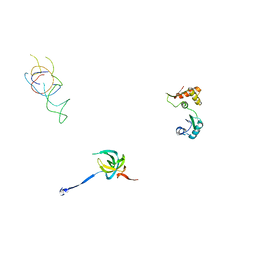 | | Coordinates of S12, L11 proteins and E-site tRNA from 70S crystal structure separately fitted into the Cryo-EM map of E.coli 70S.EF-G.GDPNP complex. The atomic coordinates originally from the E-site tRNA were fitted in the position of the hybrid P/E-site tRNA. | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, E-tRNA | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
1PN7
 
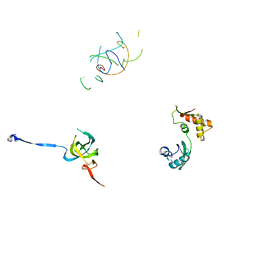 | | Coordinates of S12, L11 proteins and P-tRNA, from the 70S X-ray structure aligned to the 70S Cryo-EM map of E.coli ribosome | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, P-tRNA | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
3ZD0
 
 | | The Solution Structure of Monomeric Hepatitis C Virus p7 Yields Potent Inhibitors of Virion Release | | Descriptor: | P7 PROTEIN | | Authors: | Foster, T.L, Sthompson, G, Kalverda, A.P, Kankanala, J, Thompson, J, Barker, A.M, Clarke, D, Noerenberg, M, Pearson, A.R, Rowlands, D.J, Homans, S.W, Harris, M, Foster, R, Griffin, S.D.C. | | Deposit date: | 2012-11-23 | | Release date: | 2013-09-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-Guided Design Affirms Inhibitors of Hepatitis C Virus P7 as a Viable Class of Antivirals Targeting Virion Release
Hepatology, 59, 2014
|
|
1PN6
 
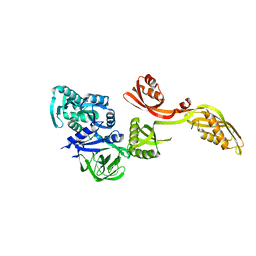 | | Domain-wise fitting of the crystal structure of T.thermophilus EF-G into the low resolution map of the release complex.Puromycin.EFG.GDPNP of E.coli 70S ribosome. | | Descriptor: | Elongation factor G | | Authors: | Valle, M, Zavialov, A, Sengupta, J, Rawat, U, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-06-12 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Locking and Unlocking of Ribosomal Motions
Cell(Cambridge,Mass.), 114, 2003
|
|
1QZC
 
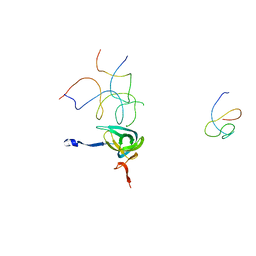 | | Coordinates of S12, SH44, LH69 and SRL separately fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S12 | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZD
 
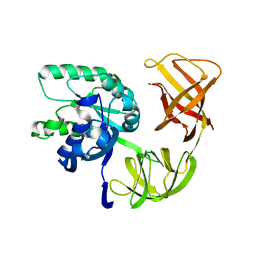 | | EF-Tu.kirromycin coordinates fitted into the cryo-EM map of EF-Tu ternary complex (GDP.Kirromycin) bound 70S ribosome | | Descriptor: | Elongation factor Tu | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1QZB
 
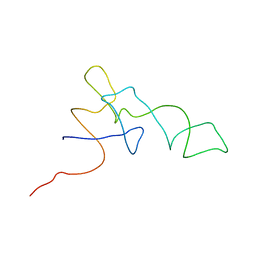 | | Coordinates of the A-site tRNA model fitted into the cryo-EM map of 70S ribosome in the pre-translocational state | | Descriptor: | Phe-tRNA | | Authors: | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | Deposit date: | 2003-09-16 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
1TVZ
 
 | | Crystal structure of 3-hydroxy-3-methylglutaryl-coenzyme A synthase from Staphylococcus aureus | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, SULFATE ION | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-06-30 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
1PL2
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 T68E mutant in complex with decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1FEU
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN TL5, ONE OF THE CTC FAMILY PROTEINS, COMPLEXED WITH A FRAGMENT OF 5S RRNA. | | Descriptor: | 19 NT FRAGMENT OF 5S RRNA, 21 NT FRAGMENT OF 5S RRNA, 50S RIBOSOMAL PROTEIN L25, ... | | Authors: | Fedorov, R.V, Meshcheryakov, V.A, Gongadze, G.M, Fomenkova, N.P, Nevskaya, N.A, Selmer, M, Laurberg, M, Kristensen, O, Al-Karadaghi, S, Liljas, A, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2000-07-23 | | Release date: | 2001-06-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ribosomal protein TL5 complexed with RNA provides new insights into the CTC family of stress proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1ZHN
 
 | | Crystal Structure of mouse CD1d bound to the self ligand phosphatidylcholine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(DODECANOYLOXY)METHYL]-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-3,5,8-TRIOXA-4-PHOSPHADOTRIACONTAN-1-AMINIUM 4-OXIDE, CD1d1 antigen, ... | | Authors: | Giabbai, B, Sidobre, S, Crispin, M.M.D, Sanchez Ruiz, Y, Bachi, A, Kronenberg, M, Wilson, I.A, Degano, M. | | Deposit date: | 2005-04-26 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation
J.Immunol., 175, 2005
|
|
1PKW
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with glutathione | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, GLUTATHIONE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1PL1
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with a decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
8B7S
 
 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
1XWG
 
 | | Human GST A1-1 T68E mutant | | Descriptor: | Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Novotny, M, Grehn, L, Olin, B, Madsen, D, Wahlberg, M, Mannervik, B, Kleywegt, G.J. | | Deposit date: | 2004-11-01 | | Release date: | 2005-11-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1PKZ
 
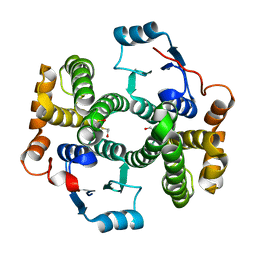 | | Crystal structure of human glutathione transferase (GST) A1-1 | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, Glutathione S-transferase A1 | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1TXT
 
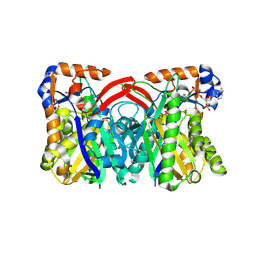 | | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase | | Descriptor: | 3-hydroxy-3-methylglutaryl-CoA synthase, ACETOACETYL-COENZYME A | | Authors: | Campobasso, N, Patel, M, Wilding, I.E, Kallender, H, Rosenberg, M, Gwynn, M. | | Deposit date: | 2004-07-06 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Staphylococcus aureus 3-hydroxy-3-methylglutaryl-CoA synthase: crystal structure and mechanism
J.Biol.Chem., 279, 2004
|
|
4XCR
 
 | | Monomeric Human Cu,Zn Superoxide dismutase, loops IV and VII deleted, apo form, mutant I35A | | Descriptor: | Superoxide dismutase [Cu-Zn] | | Authors: | Wang, H, Logan, D.T, Danielsson, J, Mu, X, Binolfi, A, Theillet, F, Bekei, B, Lang, L, Wennerstrom, H, Selenko, P, Oliveberg, M. | | Deposit date: | 2014-12-18 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.602 Å) | | Cite: | Thermodynamics of protein destabilization in live cells.
Proc. Natl. Acad. Sci. U.S.A., 112, 2015
|
|
