4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
4YY9
 
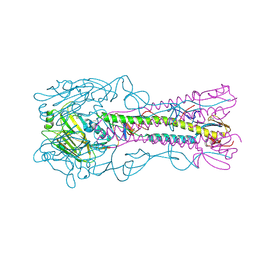 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) at 2.6 Angstroms resolution
To Be Published
|
|
4YY7
 
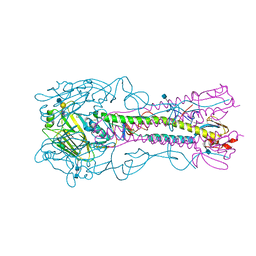 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4YY0
 
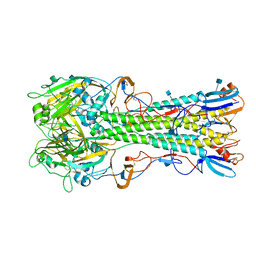 | | The structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2 | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-03-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4YYA
 
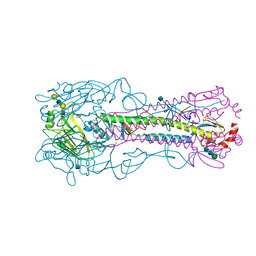 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with avian receptor analog 3'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/chicken/Taiwan/A2837/2013)
To Be Published
|
|
4YYB
 
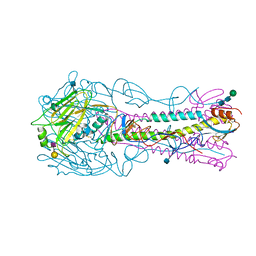 | | The structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) in complex with human receptor analog 6'SLNLN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HA1, HA2, ... | | Authors: | Wang, F, Qi, J, Bi, Y, Zhang, W, Wang, M, Wang, M, Liu, J, Yan, J, Shi, Y, Gao, G.F. | | Deposit date: | 2015-03-23 | | Release date: | 2016-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure of hemagglutinin from a H6N1 influenza virus (A/Taiwan/2/2013) at 2.6 Angstroms resolution
To Be Published
|
|
3HG1
 
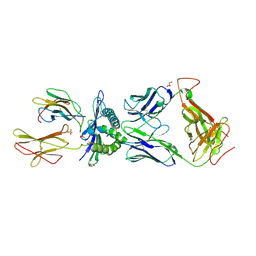 | | Germline-governed recognition of a cancer epitope by an immunodominant human T cell receptor | | Descriptor: | Beta-2-microglobulin, CANCER/MART-1, GLYCEROL, ... | | Authors: | Cole, D.K, Yuan, F, Rizkallah, P.J, Miles, J.J, Gostick, E, Price, D.A, Gao, G.F, Jakobsen, B.K, Sewell, A.K. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Germ line-governed recognition of a cancer epitope by an immunodominant human T-cell receptor.
J.Biol.Chem., 284, 2009
|
|
6KV5
 
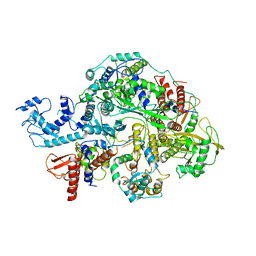 | | Structure of influenza D virus apo polymerase | | Descriptor: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
4MOD
 
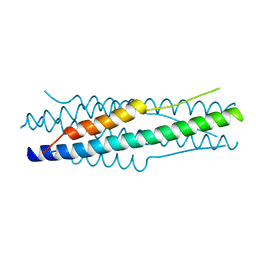 | | Structure of the MERS-CoV fusion core | | Descriptor: | HR1 of S protein, LINKER, HR2 of S protein | | Authors: | Gao, J, Lu, G, Qi, J, Li, Y, Wu, Y, Deng, Y, Geng, H, Xiao, H, Tan, W, Yan, J, Gao, G.F. | | Deposit date: | 2013-09-12 | | Release date: | 2013-10-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus.
J.Virol., 87, 2013
|
|
4MWR
 
 | | Anhui N9-zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWX
 
 | | Shanghai N9-zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWJ
 
 | | Anhui N9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWW
 
 | | Shanghai N9-oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWQ
 
 | | Anhui N9-oseltamivir carboxylate | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWL
 
 | | Shanghai N9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MX0
 
 | | Shanghai N9-peramivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWV
 
 | | Anhui N9-peramivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWU
 
 | | Anhui N9-laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
4MWY
 
 | | Shanghai N9-laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Wu, Y, Qi, J.X, Gao, F, Gao, G.F. | | Deposit date: | 2013-09-25 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of two distinct neuraminidases from avian-origin human-infecting H7N9 influenza viruses
Cell Res., 23, 2013
|
|
3TIB
 
 | | Crystal structure of 1957 pandemic H2N2 neuraminidase complexed with laninamivir octanoate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-9-O-octanoyl-D-glycero-D-galacto-non-2-enonic acid, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TI4
 
 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with laninamivir octanoate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-9-O-octanoyl-D-glycero-D-galacto-non-2-enonic acid, ACETATE ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TIA
 
 | | Crystal structure of 1957 pandemic H2N2 neuraminidase complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TIC
 
 | | Crystal structure of 1957 pandemic H2N2 neuraminidase complexed with zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TI3
 
 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with laninamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-7-O-methyl-D-glycero-D-galacto-non-2-enonic acid, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
3TI5
 
 | | Crystal structure of 2009 pandemic H1N1 neuraminidase complexed with Zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Vavricka, C.J, Li, Q, Wu, Y, Qi, J, Wang, M, Liu, Y, Gao, F, Liu, J, Feng, E, He, J, Wang, J, Liu, H, Jiang, H, Gao, G.F. | | Deposit date: | 2011-08-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional analysis of laninamivir and its octanoate prodrug reveals group specific mechanisms for influenza NA inhibition
Plos Pathog., 7, 2011
|
|
