5H5I
 
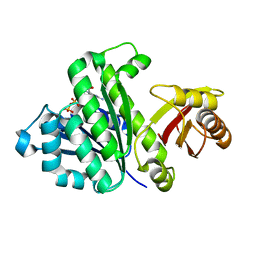 | | Staphylococcus aureus FtsZ-GDP R29A mutant in R state | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
3C5V
 
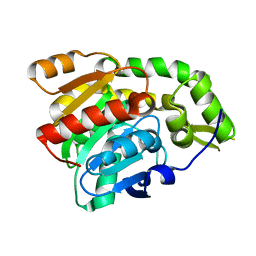 | | PP2A-specific methylesterase apo form (PME) | | Descriptor: | Protein phosphatase methylesterase 1 | | Authors: | Xing, Y, Li, Z, Chen, Y, Stock, J, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2008-02-01 | | Release date: | 2008-04-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural mechanism of demethylation and inactivation of protein phosphatase 2A.
Cell(Cambridge,Mass.), 133, 2008
|
|
5H5H
 
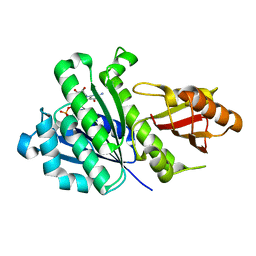 | | Staphylococcus aureus FtsZ-GDP R29A mutant in T state | | Descriptor: | CALCIUM ION, Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Fujita, J, Harada, R, Maeda, Y, Saito, Y, Mizohata, E, Inoue, T, Shigeta, Y, Matsumura, H. | | Deposit date: | 2016-11-05 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of the key interactions in structural transition pathway of FtsZ from Staphylococcus aureus
J. Struct. Biol., 198, 2017
|
|
7DHL
 
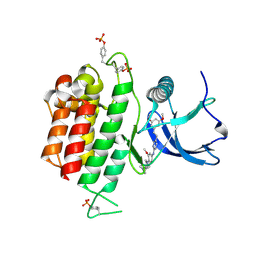 | | Crystal structure of FGFR3 in complex with pyrimidine derivative | | Descriptor: | 5-[2-(3,5-dimethoxyphenyl)ethyl]-N-[3-methoxy-4-[4-(4-methylpiperazin-1-yl)piperidin-1-yl]phenyl]pyrimidin-2-amine, Fibroblast growth factor receptor 3 | | Authors: | Echizen, Y, Tateishi, Y, Amano, Y. | | Deposit date: | 2020-11-16 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Synthesis and structure-activity relationships of pyrimidine derivatives as potent and orally active FGFR3 inhibitors with both increased systemic exposure and enhanced in vitro potency.
Bioorg.Med.Chem., 33, 2021
|
|
1B7F
 
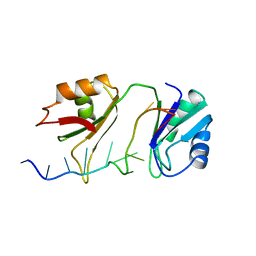 | | SXL-LETHAL PROTEIN/RNA COMPLEX | | Descriptor: | PROTEIN (SXL-LETHAL PROTEIN), RNA (5'-R(P*GP*UP*UP*GP*UP*UP*UP*UP*UP*UP*UP*U)-3') | | Authors: | Handa, N, Nureki, O, Kurimoto, K, Kim, I, Sakamoto, H, Shimura, Y, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-01-23 | | Release date: | 1999-05-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein.
Nature, 398, 1999
|
|
4KEP
 
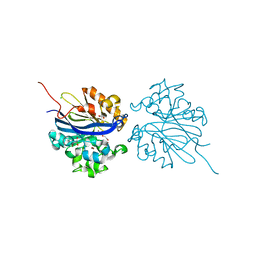 | | Crystal structure of 4-pyridoxolactonase, wild-type | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, ACETATE ION, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
4KEQ
 
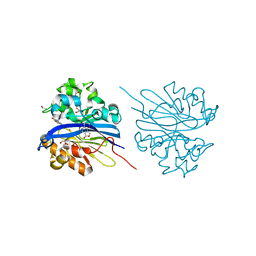 | | Crystal structure of 4-pyridoxolactonase, 5-pyridoxolactone bound | | Descriptor: | 1,2-ETHANEDIOL, 4-pyridoxolactonase, 7-hydroxy-6-methylfuro[3,4-c]pyridin-3(1H)-one, ... | | Authors: | Kobayashi, J, Yoshikane, Y, Baba, S, Mizutani, K, Takahashi, N, Mikami, B, Yagi, T. | | Deposit date: | 2013-04-26 | | Release date: | 2014-04-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.279 Å) | | Cite: | Structure of 4-pyridoxolactonase from Mesorhizobium loti.
Acta Crystallogr.,Sect.F, 70, 2014
|
|
1EOJ
 
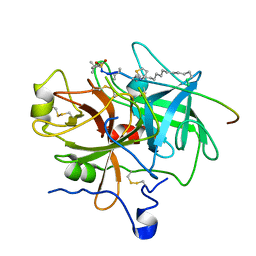 | | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures | | Descriptor: | ALPHA THROMBIN, THROMBIN INHIBITOR P798 | | Authors: | Slon-Usakiewicz, J.J, Sivaraman, J, Li, Y, Cygler, M, Konishi, Y. | | Deposit date: | 2000-03-23 | | Release date: | 2000-05-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design of P1' and P3' residues of trivalent thrombin inhibitors and their crystal structures.
Biochemistry, 39, 2000
|
|
2LHI
 
 | | Solution structure of Ca2+/CNA1 peptide-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin,Serine/threonine-protein phosphatase 2B catalytic subunit A1 | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
3VOL
 
 | | X-ray Crystal Structure of PAS-HAMP Aer2 in the CN-bound Form | | Descriptor: | Aerotaxis transducer Aer2, CYANIDE ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sawai, H, Sugimoto, H, Shiro, Y, Aono, S. | | Deposit date: | 2012-01-27 | | Release date: | 2012-05-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Structural basis for oxygen sensing and signal transduction of the heme-based sensor protein Aer2 from Pseudomonas aeruginosa
Chem.Commun.(Camb.), 48, 2012
|
|
2LHH
 
 | | Solution structure of Ca2+-bound yCaM | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Ogura, K, Takahashi, K, Kobashigawa, Y, Yoshida, R, Itoh, H, Yazawa, M, Inagaki, F. | | Deposit date: | 2011-08-10 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of yeast Saccharomyces cerevisiae calmodulin in calcium- and target peptide-bound states reveal similarities and differences to vertebrate calmodulin.
Genes Cells, 17, 2012
|
|
8WTQ
 
 | | HUMAN SQUALENE SYNTHASE IN COMPLEX WITH {1-[2-Chloro-5-(2,2-dimethyl-propyl)-13-(2-methoxy-phenyl)-6-oxo-6,7,10,11-tetrahydro-5H,9H,13H-12-oxa-5,8-diaza-benzocycloundecene-8-carbonyl]-piperidin-4-yl}-acetic acid | | Descriptor: | 2-[1-[[(10~{S})-13-chloranyl-2-(2,2-dimethylpropyl)-10-(2-methoxyphenyl)-3-oxidanylidene-9-oxa-2,5-diazabicyclo[9.4.0]pentadeca-1(15),11,13-trien-5-yl]carbonyl]piperidin-4-yl]ethanoic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Haginoya, N, Suzuki, M, Ishigai, Y, Terayama, K, Kanda, A, Sugita, K. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel 11-Membered Templates as Squalene Synthase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8WTR
 
 | | HUMAN SQUALENE SYNTHASE IN COMPLEX WITH (1S,3R)-3-[2-Chloro-5-(2,2-dimethyl-propyl)-13-(2-methoxy-phenyl)-6,8-dioxo-5,6,7,8,10,11-hexahydro-13H-12-oxa-5,9-diaza-benzocycloundecen-9-yl]-cyclohexanecarboxylic acid | | Descriptor: | (1~{S},3~{R})-3-[(10~{S})-13-chloranyl-2-(2,2-dimethylpropyl)-10-(2-methoxyphenyl)-3,5-bis(oxidanylidene)-9-oxa-2,6-diazabicyclo[9.4.0]pentadeca-1(15),11,13-trien-6-yl]cyclohexane-1-carboxylic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Haginoya, N, Suzuki, M, Ishigai, Y, Terayama, K, Kanda, A, Sugita, K. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel 11-Membered Templates as Squalene Synthase Inhibitors.
J.Med.Chem., 67, 2024
|
|
1IYO
 
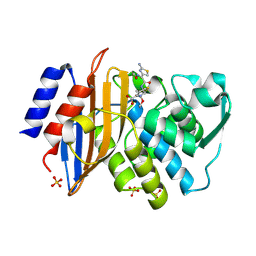 | | Toho-1 beta-Lactamase In Complex With Cefotaxime | | Descriptor: | CEFOTAXIME, C3' cleaved, open, ... | | Authors: | Shimamura, T, Ibuka, A, Fushinobu, S, Wakagi, T, Ishiguro, M, Ishii, Y, Matsuzawa, H. | | Deposit date: | 2002-09-04 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acyl-intermediate Structures of the Extended-spectrum Class A beta -Lactamase, Toho-1, in Complex with Cefotaxime, Cephalothin, and Benzylpenicillin.
J.Biol.Chem., 277, 2002
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
1UG7
 
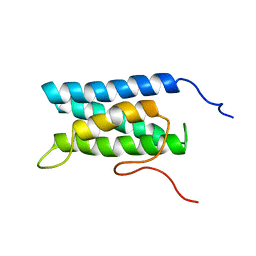 | | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806 | | Descriptor: | 2610208M17Rik protein | | Authors: | Li, H, Kigawa, T, Tomizawa, T, Koshiba, S, Inoue, M, Shirouzu, M, Terada, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-13 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of four helical up-and-down bundle domain of the hypothetical protein 2610208M17Rik similar to the protein FLJ12806
To be Published
|
|
1K2G
 
 | | Structural basis for the 3'-terminal guanosine recognition by the group I intron | | Descriptor: | 5'-R(*CP*AP*GP*AP*CP*UP*UP*CP*GP*GP*UP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*G)-3' | | Authors: | Kitamura, Y, Muto, Y, Watanabe, S, Kim, I, Ito, T, Nishiya, Y, Sakamoto, K, Ohtsuki, T, Kawai, G, Watanabe, K, Hosono, K, Takaku, H, Katoh, E, Yamazaki, T, Inoue, T, Yokoyama, S. | | Deposit date: | 2001-09-27 | | Release date: | 2002-05-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an RNA fragment with the P7/P9.0 region and the 3'-terminal guanosine of the tetrahymena group I intron.
RNA, 8, 2002
|
|
1UHS
 
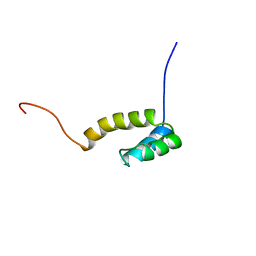 | | Solution structure of mouse homeodomain-only protein HOP | | Descriptor: | homeodomain only protein | | Authors: | Saito, K, Koshiba, S, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-10 | | Release date: | 2004-07-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mouse homeodomain-only protein HOP
To be Published
|
|
1UHR
 
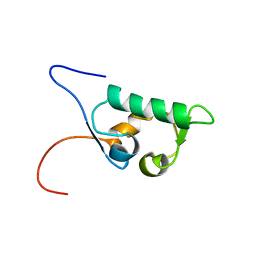 | | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a | | Descriptor: | SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily D member 1 | | Authors: | Yamada, K, Saito, K, Nameki, N, Inoue, M, Koshiba, S, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Matsuda, T, Seki, E, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-07-09 | | Release date: | 2004-08-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SWIB domain of mouse BRG1-associated factor 60a
To be Published
|
|
7COY
 
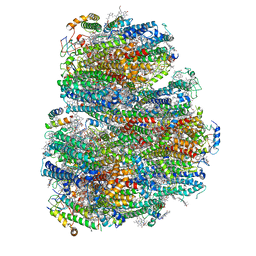 | | Structure of the far-red light utilizing photosystem I of Acaryochloris marina | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL D, ... | | Authors: | Kawakami, K, Yonekura, K, Hamaguchi, T, Kashino, Y, Shinzawa-Itoh, K, Inoue-Kashino, N, Itoh, S, Ifuku, K, Yamashita, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-03-31 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the far-red light utilizing photosystem I of Acaryochloris marina.
Nat Commun, 12, 2021
|
|
3BOE
 
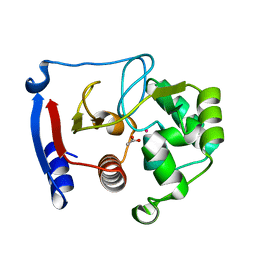 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 2 with acetate (CDCA1-R2) | | Descriptor: | ACETATE ION, CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
3BOJ
 
 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 1 without bound metal (CDCA1-R1) | | Descriptor: | ACETATE ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
3BOB
 
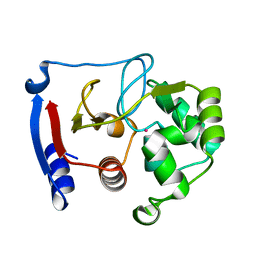 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 2 | | Descriptor: | CADMIUM ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
3BOC
 
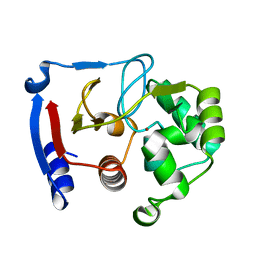 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- zinc bound domain 2 (CDCA1-R2) | | Descriptor: | Cadmium-specific carbonic anhydrase, ZINC ION | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
1AF3
 
 | | RAT BCL-XL AN APOPTOSIS INHIBITORY PROTEIN | | Descriptor: | APOPTOSIS REGULATOR BCL-X | | Authors: | Aritomi, M, Kunishima, N, Inohara, N, Ishibashi, Y, Ohta, S, Morikawa, K. | | Deposit date: | 1997-03-21 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat Bcl-xL. Implications for the function of the Bcl-2 protein family.
J.Biol.Chem., 272, 1997
|
|
