1RE5
 
 | | Crystal structure of 3-carboxy-cis,cis-muconate lactonizing enzyme from Pseudomonas putida | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3-carboxy-cis,cis-muconate cycloisomerase, CITRIC ACID | | Authors: | Yang, J, Wang, Y, Woolridge, E.M, Petsko, G.A, Kozarich, J.W, Ringe, D. | | Deposit date: | 2003-11-06 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of 3-Carboxy-cis,cis-muconate Lactonizing Enzyme from Pseudomonas putida, a Fumarase Class II Type Cycloisomerase: Enzyme Evolution in Parallel Pathways.
Biochemistry, 43, 2004
|
|
5KCD
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-methyl Substituted OBHS-N derivative | | Descriptor: | (1S,2R,4S)-5,6-bis(4-hydroxyphenyl)-N-methyl-N-phenyl-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
1NQP
 
 | | Crystal structure of Human hemoglobin E at 1.73 A resolution | | Descriptor: | CYANIDE ION, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Dasgupta, J, Sen, U, Choudhury, D, Dutta, P, Basu, S, Chakrabarti, A, Chakrabarty, A, Dattagupta, J.K. | | Deposit date: | 2003-01-22 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystallization and preliminary X-ray structural Studies of Hemoglobin A2 and Hemoglobin E, isolated from the blood samples of Beta-thalassemic patients
Biochem.Biophys.Res.Commun., 303, 2004
|
|
1RGI
 
 | | Crystal structure of gelsolin domains G1-G3 bound to actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Burtnick, L.D, Urosev, D, Irobi, E, Narayan, K, Robinson, R.C. | | Deposit date: | 2003-11-12 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF
Embo J., 23, 2004
|
|
5KGF
 
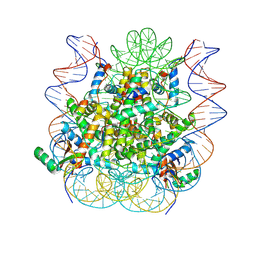 | | Structural model of 53BP1 bound to a ubiquitylated and methylated nucleosome, at 4.5 A resolution | | Descriptor: | DNA (145-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Wilson, M.D, Benlekbir, S, Sicheri, F, Rubinstein, J.L, Durocher, D. | | Deposit date: | 2016-06-13 | | Release date: | 2016-07-27 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | The structural basis of modified nucleosome recognition by 53BP1.
Nature, 536, 2016
|
|
2R72
 
 | | Crystal structure of infectious bursal disease virus VP1 polymerase, incubated with Mg2+ ion. | | Descriptor: | INFECTIOUS BURSAL DISEASE VIRUS VP1 POLYMERASE, MAGNESIUM ION | | Authors: | Garriga, D, Navarro, A, Querol-Audi, J, Abaitua, F, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-09-07 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Activation mechanism of a noncanonical RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1NN0
 
 | | Crystal structure of human thymidylate kinase with ddTMP and ADP | | Descriptor: | 3'-DEOXYTHYMIDINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ostermann, N, Segura-Pena, D, Meier, C, Veit, T, Monnerjahn, M, Konrad, M, Lavie, A. | | Deposit date: | 2003-01-12 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of human thymidylate kinase in complex with prodrugs:
implications for the structure-based design of novel compounds
Biochemistry, 42, 2003
|
|
5KCF
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-methoxybenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-ethyl-5,6-bis(4-hydroxyphenyl)-N-(4-methoxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-06 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
5KCT
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with an N-ethyl, 4-chlorobenzyl OBHS-N derivative | | Descriptor: | (1R,2S,4R)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, (1S,2R,4S)-N-(4-chlorophenyl)-N-ethyl-5,6-bis(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-5-ene-2-sulfonamide, Estrogen receptor, ... | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Dharmarajan, V, Goswami, D, Kastrati, I, Novick, S, Nowak, J, Zhou, H.B, Boonmuen, N, Zhao, Y, Min, J, Frasor, J, Katzenellenbogen, B.S, Griffin, P.R, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-06-07 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Full antagonism of the estrogen receptor without a prototypical ligand side chain.
Nat. Chem. Biol., 13, 2017
|
|
2R9M
 
 | | Cathepsin S complexed with Compound 15 | | Descriptor: | Cathepsin S, N-[(1S)-2-[(4-cyano-1-methylpiperidin-4-yl)amino]-1-(cyclohexylmethyl)-2-oxoethyl]morpholine-4-carboxamide | | Authors: | Ward, Y.D, Emmanuel, M.J, Thomson, D.S, Liu, W, Bekkali, Y, Frye, L.L, Girardot, M, Morwick, T, Young, E.R.R, Zindell, R, Hrapchak, M, DeTuri, M, White, A, Crane, K.M, White, D.M, Wang, Y, Hao, M.-H, Grygon, C.A, Labadia, M.E, Wildeson, J, Freeman, D, Nelson, R, Capolino, A, Peterson, J.D, Raymond, E.L, Brown, M.L, Spero, D.M. | | Deposit date: | 2007-09-13 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Design and Synthesis of Reversible Inhibitors of Cathepsin S: alpha,alpha-Disubstitution at the P1 Residue Provides Potent Inhibitors in Cellular Assays and In Vivo Models of Antigen Presentation
To be Published
|
|
1RQL
 
 | | Crystal Structure of Phosponoacetaldehyde Hydrolase Complexed with Magnesium and the Inhibitor Vinyl Sulfonate | | Descriptor: | MAGNESIUM ION, Phosphonoacetaldehyde Hydrolase, VINYLSULPHONIC ACID | | Authors: | Morais, M.C, Zhang, G, Zhang, W, Olsen, D.B, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2003-12-05 | | Release date: | 2004-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic and site-directed mutagenesis
analysis of the mechanism of Schiff-base formation in
phosphonoacetaldehyde hydrolase catalysis
J.Biol.Chem., 279, 2004
|
|
1NXM
 
 | | The high resolution structures of RmlC from Streptococcus suis | | Descriptor: | dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase | | Authors: | Dong, C, Major, L.L, Allen, A, Blankenfeldt, W, Maskell, D, Naismith, J.H. | | Deposit date: | 2003-02-11 | | Release date: | 2003-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-Resolution Structures of RmlC from Streptococcus suis in Complex with Substrate Analogs Locate the Active Site of This Class of Enzyme
Structure, 11, 2003
|
|
7NHG
 
 | |
7NIA
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ059a | | Descriptor: | 4-[[(3R)-3-cyclopropyl-2-azaspiro[3.3]heptan-2-yl]methyl]-N-[[(3R)-3-oxidanyl-1-[6-[(phenylmethyl)amino]pyrimidin-4-yl]piperidin-3-yl]methyl]benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-02-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
1NYR
 
 | | Structure of Staphylococcus aureus threonyl-tRNA synthetase complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, THREONINE, ZINC ION, ... | | Authors: | Torres-Larios, A, Sankaranarayanan, R, Rees, B, Dock-Bregeon, A.C, Moras, D. | | Deposit date: | 2003-02-13 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational movements and cooperativity upon amino acid, ATP and tRNA binding in threonyl-tRNA synthetase
J.Mol.Biol., 331, 2003
|
|
7NI9
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ058 | | Descriptor: | ACETATE ION, N-(((R)-1-(6-(benzylamino)pyrimidin-4-yl)-3-hydroxypiperidin-3-yl)methyl)-4-(((S)-1-isopropyl-2-azaspiro[3.3]heptan-2-yl)methyl)benzamide, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-02-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
1R2B
 
 | | Crystal structure of the BCL6 BTB domain complexed with a SMRT co-repressor peptide | | Descriptor: | B-cell lymphoma 6 protein, Nuclear receptor co-repressor 2 | | Authors: | Ahmad, K.F, Melnick, A, Lax, S.A, Bouchard, D, Liu, J, Kiang, C.L, Mayer, S, Licht, J.D, Prive, G.G. | | Deposit date: | 2003-09-26 | | Release date: | 2003-12-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain.
Mol.Cell, 12, 2003
|
|
7NHV
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ016 | | Descriptor: | (R)-N-((1-(6-(benzylamino)pyrimidin-4-yl)-3-hydroxypiperidin-3-yl)methyl)-4-((4,4-dimethylpiperidin-1-yl)methyl)benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-02-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7NHH
 
 | |
5RHD
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with SF013 (Mpro-x2193) | | Descriptor: | 1-[4-(methylsulfonyl)phenyl]piperazine, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, Keeley, A, Keseru, G.M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-16 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
7NI7
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ031 | | Descriptor: | (R)-4-((4,4-dimethylpiperidin-1-yl)methyl)-2-hydroxy-N-((3-hydroxy-1-(6-((3-(methylcarbamoyl)benzyl)amino)pyrimidin-4-yl)piperidin-3-yl)methyl)benzamide, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-02-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7NID
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ078 | | Descriptor: | (R)-1-(6-(benzylamino)pyrimidin-4-yl)-3-(((6-((4,4-dimethylpiperidin-1-yl)methyl)naphthalen-1-yl)amino)methyl)piperidin-3-ol, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-02-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
7NHJ
 
 | |
2RC4
 
 | | Crystal Structure of the HAT domain of the human MOZ protein | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase MYST3, ZINC ION | | Authors: | Holbert, M.A, Sikorski, T, Snowflack, D, Marmorstein, R. | | Deposit date: | 2007-09-19 | | Release date: | 2007-11-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human monocytic leukemia zinc finger histone acetyltransferase domain contains DNA-binding activity implicated in chromatin targeting.
J.Biol.Chem., 282, 2007
|
|
7NI8
 
 | | Crystal structure of the human METTL3-METTL14 complex with compound UOZ040a | | Descriptor: | (R)-4-((2-azaspiro[3.3]heptan-2-yl)methyl)-N-((1-(6-(benzylamino)pyrimidin-4-yl)-3-hydroxypiperidin-3-yl)methyl)benzamide, ACETATE ION, N6-adenosine-methyltransferase catalytic subunit, ... | | Authors: | Bedi, R.K, Huang, D, Caflisch, A. | | Deposit date: | 2021-02-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Design of Inhibitors of the m6A-RNA Writer Enzyme METTL3
Acs Bio Med Chem Au, 2023
|
|
