3BRO
 
 | | Crystal structure of the transcription regulator MarR from Oenococcus oeni PSU-1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Transcriptional regulator | | Authors: | Kim, Y, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal Structure of the Transcription Regulator MarR from Oenococcus oeni PSU-1.
To be Published
|
|
3BV6
 
 | | Crystal structure of uncharacterized metallo protein from Vibrio cholerae with beta-lactamase like fold | | Descriptor: | FE (III) ION, Metal-dependent hydrolase | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Yang, X, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of uncharacterized metallo protein from Vibrio cholerae with beta-lactamase like fold.
To be Published
|
|
1XV2
 
 | | Crystal Structure of a Protein of Unknown Function Similar to Alpha-acetolactate Decarboxylase from Staphylococcus aureus | | Descriptor: | ZINC ION, hypothetical protein, similar to alpha-acetolactate decarboxylase | | Authors: | Duke, N.E.C, Zhang, R, Quartey, P, Collart, F, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a hypothetical protein similar to alpha-acetolactate
To be Published
|
|
3CO5
 
 | | Crystal structure of sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae | | Descriptor: | BETA-MERCAPTOETHANOL, Putative two-component system transcriptional response regulator | | Authors: | Osipiuk, J, Hendricks, R, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-27 | | Release date: | 2008-04-08 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of Sigma-54 interaction domain of putative transcriptional response regulator from Neisseria gonorrhoeae.
To be Published
|
|
3CQY
 
 | | Crystal structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1 | | Descriptor: | Anhydro-N-acetylmuramic acid kinase, CHLORIDE ION, SUCCINIC ACID | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-03 | | Release date: | 2008-04-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of a functionally unknown protein (SO_1313) from Shewanella oneidensis MR-1.
To be Published
|
|
3CJ8
 
 | | Crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Enterococcus faecalis V583 | | Descriptor: | 2,3,4,5-tetrahydropyridine-2,6-dicarboxylate N-acetyltransferase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase from Enterococcus faecalis V583.
To be Published
|
|
3CNV
 
 | | Crystal structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica | | Descriptor: | CHLORIDE ION, CITRATE ANION, Putative GntR-family transcriptional regulator | | Authors: | Zimmerman, M.D, Xu, X, Cui, H, Filippova, E.V, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-26 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the ligand-binding domain of a putative GntR-family transcriptional regulator from Bordetella bronchiseptica.
To be Published
|
|
3RAG
 
 | |
3CWF
 
 | | Crystal structure of PAS domain of two-component sensor histidine kinase | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alkaline phosphatase synthesis sensor protein phoR | | Authors: | Chang, C, Tesar, C, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-21 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Extracytoplasmic PAS-like domains are common in signal transduction proteins.
J.Bacteriol., 192, 2010
|
|
3D0K
 
 | | Crystal structure of the LpqC, poly(3-hydroxybutyrate) depolymerase from Bordetella parapertussis | | Descriptor: | CHLORIDE ION, FORMIC ACID, Putative poly(3-hydroxybutyrate) depolymerase LpqC, ... | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-01 | | Release date: | 2008-07-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of the LpqC, Poly(3-hydroxybutyrate) Depolymerase from Bordetella parapertussis.
To be Published
|
|
1Y9W
 
 | | Structural Genomics, 1.9A crystal structure of an acetyltransferase from Bacillus cereus ATCC 14579 | | Descriptor: | Acetyltransferase | | Authors: | Zhang, R, Li, H, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-16 | | Release date: | 2005-02-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9A crystal structure of an acetyltransferase from Bacillus cereus ATCC 14579
To be Published, 2005
|
|
3D1R
 
 | | Structure of E. coli GlpX with its substrate fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Singer, A, Skarina, T, Dong, A, Brown, G, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
3CNE
 
 | | Crystal structure of the putative protease I from Bacteroides thetaiotaomicron | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, Putative protease I, ... | | Authors: | Zhang, R, Volkart, L, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The crystal structure of the putative protease I from Bacteroides thetaiotaomicron.
To be Published
|
|
3CZQ
 
 | | Crystal structure of putative polyphosphate kinase 2 from Sinorhizobium meliloti | | Descriptor: | FORMIC ACID, GLYCEROL, Putative polyphosphate kinase 2 | | Authors: | Osipiuk, J, Evdokimova, E, Nocek, B, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Polyphosphate-dependent synthesis of ATP and ADP by the family-2 polyphosphate kinases in bacteria.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3D6K
 
 | | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Putative aminotransferase, ... | | Authors: | Tan, K, Zhang, R, Duggan, E, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-19 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of a putative aminotransferase from Corynebacterium diphtheriae
To be Published
|
|
3D7L
 
 | |
2KKX
 
 | | Solution Structure of C-terminal domain of reduced NleG2-3 (residues 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | Descriptor: | Uncharacterized protein ECs2156 | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
3D6W
 
 | | LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus. | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-20 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of LytTr DNA-binding domain of putative methyl-accepting/DNA response regulator from Bacillus cereus.
To be Published
|
|
3CED
 
 | | Crystal structure of the C-terminal NIL domain of an ABC transporter protein homologue from Staphylococcus aureus | | Descriptor: | 1,4-BUTANEDIOL, Methionine import ATP-binding protein metN 2 | | Authors: | Cuff, M.E, Bigelow, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-28 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structure of the C-terminal NIL domain of an ABC transporter protein homologue from Staphylococcus aureus.
TO BE PUBLISHED
|
|
3CIT
 
 | | Crystal structure of the GAF domain of a putative sensor histidine kinase from Pseudomonas syringae pv. tomato | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Cuff, M.E, Li, H, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-11 | | Release date: | 2008-05-13 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the GAF domain of a putative sensor histidine kinase from Pseudomonas syringae pv. tomato
To be Published
|
|
3CNG
 
 | | Crystal structure of NUDIX hydrolase from Nitrosomonas europaea | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Osipiuk, J, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystal structure of NUDIX hydrolase from Nitrosomonas europaea.
To be Published
|
|
3U1D
 
 | | The structure of a protein with a GntR superfamily winged-helix-turn-helix domain from Halomicrobium mukohataei. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Cuff, M.E, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-12-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a protein with a GntR superfamily winged-helix-turn-helix domain from Halomicrobium mukohataei.
TO BE PUBLISHED
|
|
3GJY
 
 | |
3TX6
 
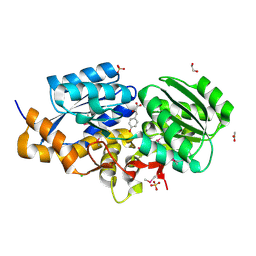 | | The Structure of a putative ABC-transporter periplasmic component from Rhodopseudomonas palustris | | Descriptor: | 1,2-ETHANEDIOL, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ACETIC ACID, ... | | Authors: | Cuff, M.E, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-22 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3KD8
 
 | |
