7RQC
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MFI-tripeptidyl-tRNA analog ACCA-IFM at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5AR8
 
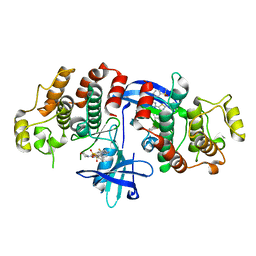 | | RIP2 Kinase Catalytic Domain (1 - 310) complex with Biphenylsulfonamide | | Descriptor: | 2,6-bis(fluoranyl)-N-[3-[5-[2-[(3-methylsulfonylphenyl)amino]pyrimidin-4-yl]-2-morpholin-4-yl-1,3-thiazol-4-yl]phenyl]benzenesulfonamide, RECEPTOR-INTERACTING SERINE/THREONINE-PROTEIN KINASE 2 | | Authors: | Charnley, A.K, Convery, M.A, Lakdawala Shah, A, Jones, E, Hardwicke, P, Bridges, A, Votta, B.J, Gough, P.J, Marquis, R.W, Bertin, J, Casillas, L. | | Deposit date: | 2015-09-24 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal Structures of Human Rip2 Kinase Catalytic Domain Complexed with ATP-Competitive Inhibitors: Foundations for Understanding Inhibitor Selectivity.
Bioorg.Med.Chem., 23, 2015
|
|
1XXF
 
 | | Crystal Structure of the FXIa Catalytic Domain in Complex with Ecotin Mutant (EcotinP) | | Descriptor: | Coagulation factor XI, Ecotin, SODIUM ION | | Authors: | Jin, L, Pandey, P, Babine, R.E, Gorga, J.C, Seidl, K.J, Gelfand, E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2004-11-04 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structures of the FXIa Catalytic Domain in Complex with Ecotin Mutants Reveal Substrate-like Interactions
J.Biol.Chem., 280, 2005
|
|
1XUP
 
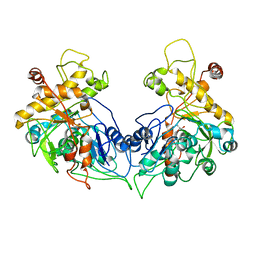 | | ENTEROCOCCUS CASSELIFLAVUS GLYCEROL KINASE COMPLEXED WITH GLYCEROL | | Descriptor: | GLYCEROL, Glycerol kinase | | Authors: | Yeh, J.I, Charrier, V, Paulo, J, Hou, L, Darbon, E, Hol, W.G.J, Deutscher, J. | | Deposit date: | 2004-10-26 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of Enterococcal Glycerol Kinase in the Absence and Presence of Glycerol: Correlation of Conformation to Substrate Binding and a Mechanism of Activation by Phosphorylation
Biochemistry, 43, 2004
|
|
1XY5
 
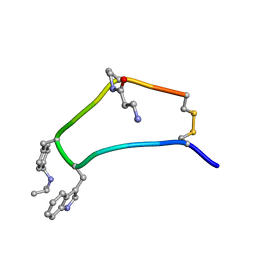 | | NMR strcutre of sst1-selective somatostatin (SRIF) analog 1 | | Descriptor: | SST1-selective somatosatin analog | | Authors: | Grace, C.R.R, Durrer, L, Koerber, S.C, Erchegyi, J, Reubi, J.C, Rivier, J.E, Riek, R. | | Deposit date: | 2004-11-09 | | Release date: | 2005-02-15 | | Last modified: | 2011-09-28 | | Method: | SOLUTION NMR | | Cite: | Somatostatin receptor 1 selective analogues: 4. Three-dimensional consensus structure by NMR
J.Med.Chem., 48, 2005
|
|
7R2V
 
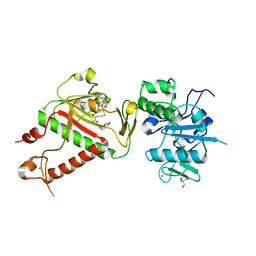 | | Structure of nsp14 from SARS-CoV-2 in complex with SAH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, Proofreading exoribonuclease nsp14, ... | | Authors: | Czarna, A, Plewka, J, Kresik, L, Matsuda, A, Abdulkarim, K, Robinson, C, OByrne, S, Cunningham, F, Georgiou, I, Pachota, M, Popowicz, G.M, Wyatt, P.G, Dubin, G, Pyrc, K. | | Deposit date: | 2022-02-06 | | Release date: | 2022-03-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Refolding of lid subdomain of SARS-CoV-2 nsp14 upon nsp10 interaction releases exonuclease activity.
Structure, 30, 2022
|
|
5AM7
 
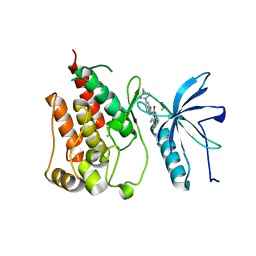 | | FGFR1 mutant with an inhibitor | | Descriptor: | 4-amino-5-fluoro-3-[5-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]quinolin-2(1H)-one, CHLORIDE ION, FIBROBLAST GROWTH FACTOR RECEPTOR 1 | | Authors: | Bunney, T.D, Wan, S, Thiyagarajan, N, Sutto, L, Williams, S.V, Ashford, P, Koss, H, Knowles, M.A, Gervasio, F.L, Coveney, P.V, Katan, M. | | Deposit date: | 2015-03-10 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | The Effect of Mutations on Drug Sensitivity and Kinase Activity of Fibroblast Growth Factor Receptors: A Combined Experimental and Theoretical Study
Ebiomedicine, 2, 2015
|
|
1XYH
 
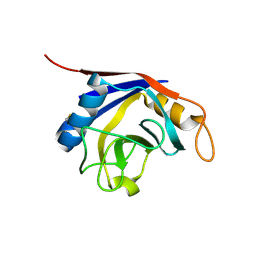 | | Crystal Structure of Recombinant Human Cyclophilin J | | Descriptor: | cyclophilin-like protein PPIL3b | | Authors: | Huang, L.-L, Zhao, X.-M, Huang, C.-Q, Yu, L, Xia, Z.-X. | | Deposit date: | 2004-11-10 | | Release date: | 2005-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of recombinant human cyclophilin J, a novel member of the cyclophilin family.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5AN9
 
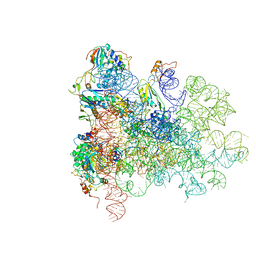 | | Mechanism of eIF6 release from the nascent 60S ribosomal subunit | | Descriptor: | 26S RIBOSOMAL RNA, 60S ACIDIC RIBOSOMAL PROTEIN P0, 60S RIBOSOMAL PROTEIN L10, ... | | Authors: | Weis, F, Giudice, E, Churcher, M, Jin, L, Hilcenko, C, Wong, C.C, Traynor, D, Kay, R.R, Warren, A.J. | | Deposit date: | 2015-09-06 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of Eif6 Release from the Nascent 60S Ribosomal Subunit
Nat.Struct.Mol.Biol., 22, 2015
|
|
4GBQ
 
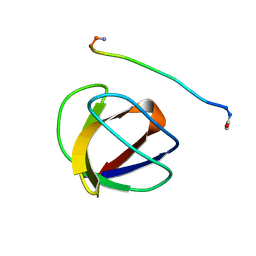 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
5Z4C
 
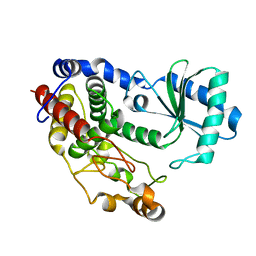 | | Crystal structure of Tailor | | Descriptor: | Terminal uridylyltransferase Tailor | | Authors: | Cheng, L, Li, F, Jiang, Y, Yu, H, Xie, C, Shi, Y, Gong, Q. | | Deposit date: | 2018-01-10 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into a unique preference for 3' terminal guanine of mirtron in Drosophila TUTase tailor.
Nucleic Acids Res., 47, 2019
|
|
6JQM
 
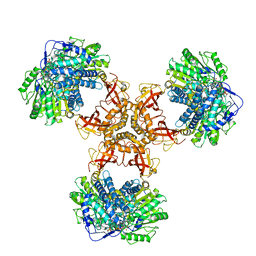 | | Structure of PaaZ with NADPH | | Descriptor: | Bifunctional protein PaaZ, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gakher, L, Vinothkumar, K.R, Katagihallimath, N, Sowdhamini, R, Sathyanarayanan, N, Cannone, G. | | Deposit date: | 2019-03-31 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis for metabolite channeling in a ring opening enzyme of the phenylacetate degradation pathway.
Nat Commun, 10, 2019
|
|
5MI0
 
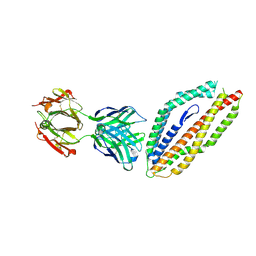 | | A thermally stabilised version of Plasmodium falciparum RH5 | | Descriptor: | MONOCLONAL ANTIBODY 9AD4, Reticulocyte binding-like protein 5,Reticulocyte binding protein 5 | | Authors: | Campeotto, I, Goldenzweig, A, Davey, J, Barfod, L, Marshall, J.M, Silk, S.E, Wright, K.E, Draper, S.J, Higgins, M.K, Fleishman, S.J. | | Deposit date: | 2016-11-27 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | One-step design of a stable variant of the malaria invasion protein RH5 for use as a vaccine immunogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7RQB
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MAI-tripeptidyl-tRNA analog ACCA-IAM at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1N5G
 
 | |
5Z87
 
 | | Structural of a novel b-glucosidase EmGH1 at 2.3 angstrom from Erythrobacter marinus | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, BENZAMIDINE, ... | | Authors: | Li, J.X, Hu, X.J, Zhao, Y, Li, L. | | Deposit date: | 2018-01-31 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical analysis of a novel b-glucosidase EmGH1 from Erythrobacter marinus
To Be Published
|
|
7RGF
 
 | | Protocadherin gammaC4 EC1-4 crystal structure disrupted trans interface | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2021-07-15 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | How clustered protocadherin binding specificity is tuned for neuronal self-/nonself-recognition.
Elife, 11, 2022
|
|
1ZHR
 
 | | Crystal Structure of the Catalytic Domain of Coagulation Factor XI in Complex with Benzamidine (S434A-T475A-C482S-K437A Mutant) | | Descriptor: | BENZAMIDINE, coagulation factor XI | | Authors: | Jin, L, Pandey, P, Babine, R.E, Weaver, D.T, Abdel-Meguid, S.S, Strickler, J.E. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: an essential step for the iterative structure-based design of factor XI inhibitors.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1Z2Z
 
 | | Crystal Structure of the Putative tRNA pseudouridine synthase D (TruD) from Methanosarcina mazei, Northeast Structural Genomics Target MaR1 | | Descriptor: | Probable tRNA pseudouridine synthase D | | Authors: | Forouhar, F, Yong, W, Ciano, M, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-03-10 | | Release date: | 2005-04-26 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Putative tRNA pseudouridine synthase D (TruD) from Methanosarcina mazei, Northeast Structural Genomics Target MaR1
To be Published
|
|
5WEX
 
 | | Discovery of new selenoureido analogs of 4-(4-fluorophenylureido) benzenesulfonamides as carbonic anhydrase inhibitors | | Descriptor: | 4-{[(4-fluorophenyl)carbamothioyl]amino}benzene-1-sulfonamide, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Angeli, A, Tanini, D, Peat, T.S, Di Cesare Mannelli, L, Bartolucci, G, Capperucci, A, Ghelardini, C, Supuran, C.T, Carta, F. | | Deposit date: | 2017-07-10 | | Release date: | 2017-10-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of New Selenoureido Analogues of 4-(4-Fluorophenylureido)benzenesulfonamide as Carbonic Anhydrase Inhibitors.
ACS Med Chem Lett, 8, 2017
|
|
5MOB
 
 | | ABA RECEPTOR FROM TOMATO, SlPYL1 | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, SULFATE ION, SlPYL1_ABA | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.669 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
5W8R
 
 | | Toxoplasma Gondii CDPK1 in complex with inhibitor 3CIB-PPI | | Descriptor: | 1-tert-butyl-3-[(3-chlorophenyl)methyl]-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALCIUM ION, Calmodulin-domain protein kinase 1 | | Authors: | El Bakkouri, M, Lovato, D, Loppnau, P, Lin, Y.H, Rutaganaria, F, Lopez, M.S, Shokat, L, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Sibley, D, Hui, R, Walker, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-22 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toxoplasma Gondii CDPK1 in complex with inhibitor 3CIB-PPI
To be published
|
|
5WAX
 
 | | Crystal Structure of Sugarcane SAPK10 (serine/threonine-protein kinase 10) | | Descriptor: | GLYCEROL, SAPK10 (serine/threonine-protein kinase 10) | | Authors: | Righetto, G.L, Counago, R.M, Halabelian, L, Santiago, A.S, Massirer, K.B, Arruda, P, Gileadi, O, Menossi, M, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-27 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Sugarcane SAPK10 (serine/threonine-protein kinase 10)
To Be Published
|
|
1N65
 
 | |
1N6J
 
 | | Structural basis of sequence-specific recruitment of histone deacetylases by Myocyte Enhancer Factor-2 | | Descriptor: | 5'-D(*AP*GP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*GP*C)-3', 5'-D(*GP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*CP*T)-3', Calcineurin-binding protein Cabin 1, ... | | Authors: | Han, A, Pan, F, Stroud, J.C, Youn, H.D, Liu, J.O, Chen, L. | | Deposit date: | 2002-11-11 | | Release date: | 2003-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2
Nature, 422, 2003
|
|
